Hedgehog signaling pathway Indian hedgehog expressed in gut



- Slides: 24

Hedgehog signaling pathway • Indian hedgehog: expressed in gut and chondrocytes • Desert hedgehog: expressed in sertoli cells of the testes • Sonic hedgehog: Involved in many developmental processes. Best characterized

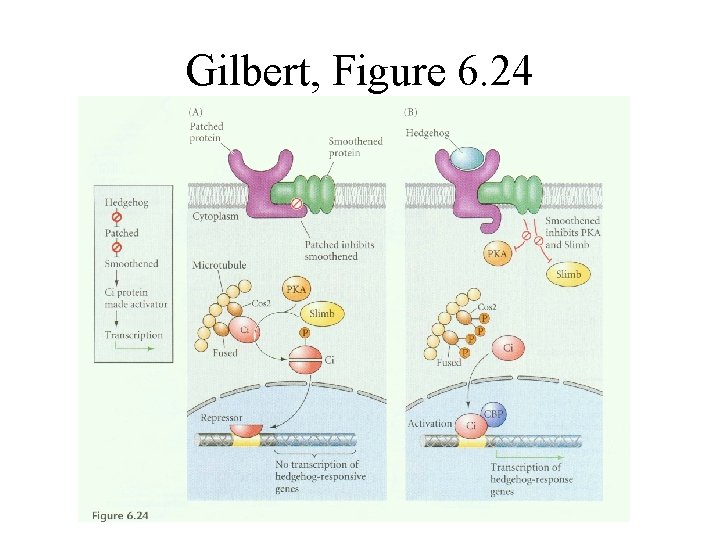
Gilbert, Figure 6. 24

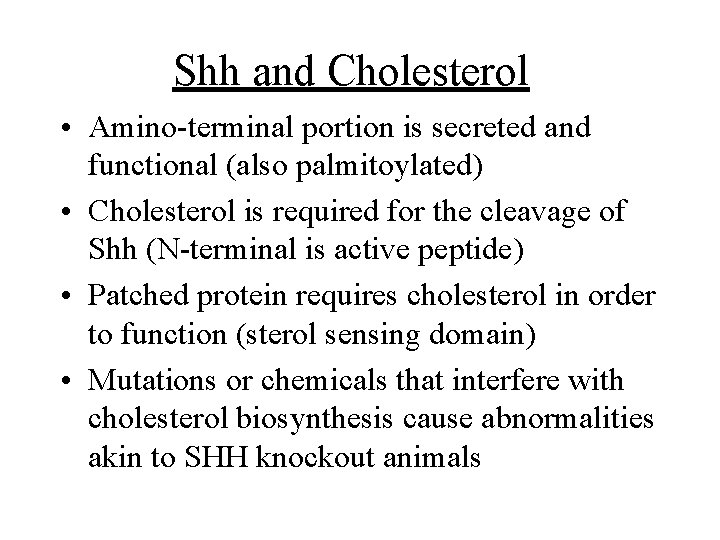
Shh and Cholesterol • Amino-terminal portion is secreted and functional (also palmitoylated) • Cholesterol is required for the cleavage of Shh (N-terminal is active peptide) • Patched protein requires cholesterol in order to function (sterol sensing domain) • Mutations or chemicals that interfere with cholesterol biosynthesis cause abnormalities akin to SHH knockout animals

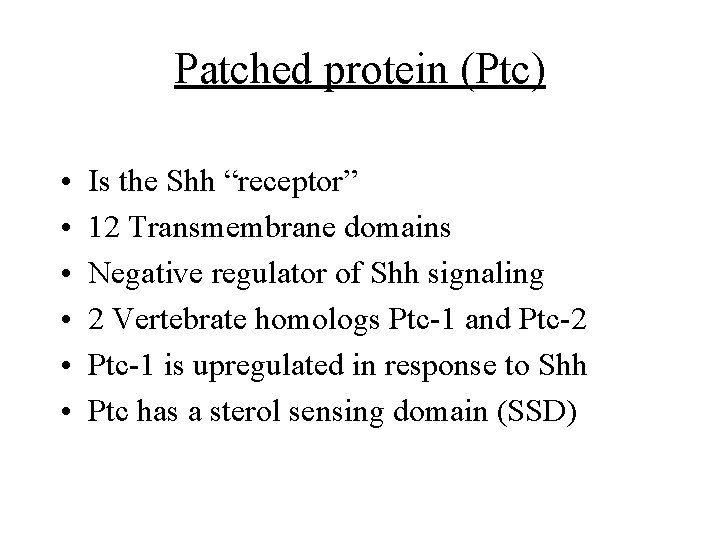
Patched protein (Ptc) • • • Is the Shh “receptor” 12 Transmembrane domains Negative regulator of Shh signaling 2 Vertebrate homologs Ptc-1 and Ptc-2 Ptc-1 is upregulated in response to Shh Ptc has a sterol sensing domain (SSD)

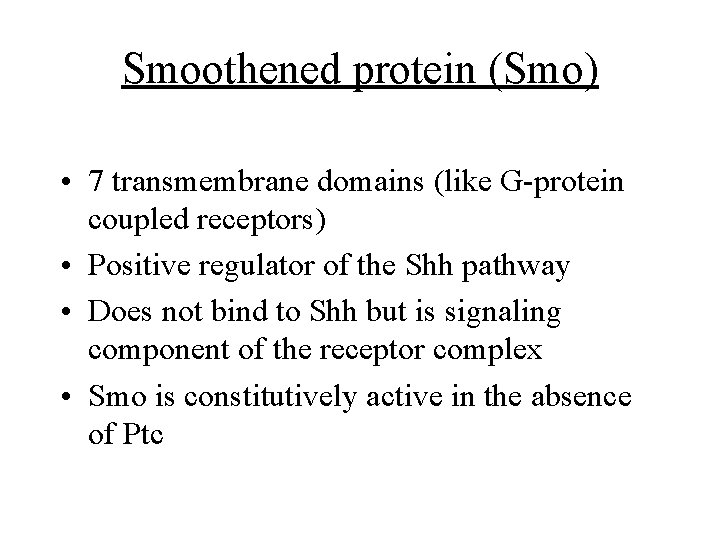
Smoothened protein (Smo) • 7 transmembrane domains (like G-protein coupled receptors) • Positive regulator of the Shh pathway • Does not bind to Shh but is signaling component of the receptor complex • Smo is constitutively active in the absence of Ptc

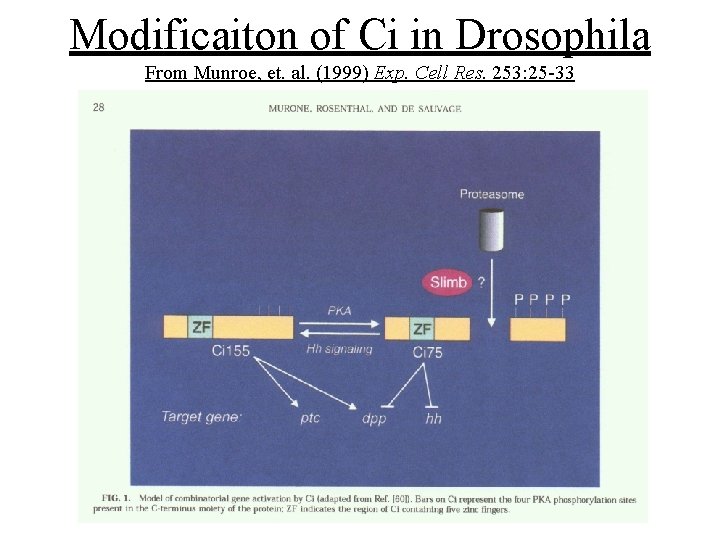
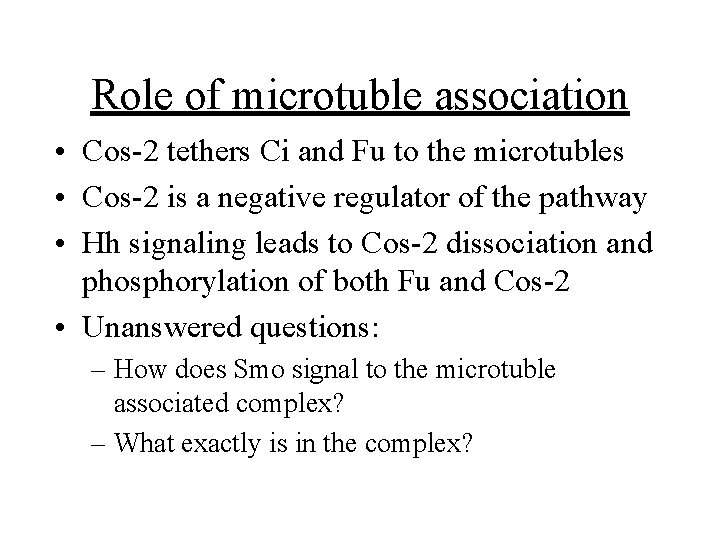
Cubitus interruptus (Ci)/Gli transcription factor family • Zn++ finger transcription factors • Recognize a 9 bp consensus sequence in the promoters of several Hh target genes • In the absense of Hh the full length protein (Ci-155) is in the cytoplasm and gets processed into a 75 k. D N-terminal repressor form (Ci-75)

Modificaiton of Ci in Drosophila From Munroe, et. al. (1999) Exp. Cell Res. 253: 25 -33

Ci-75 repressor generation is: • Triggered in part by protein kinase A (Pka) phosphorylation of the C-terminus • Possible participation of Slimb, a ubiquitin targeting protein and the proteosome • Cleavage of Ci-155 is blocked by the presence of Hedgehog (Smo signaling)

Hedgehog activation of “positively regulating” Ci-155 • Facilitated by a serine/threonine kinase called fused • Antagonized by “Supressor of fused” Su(fu) • Mutant embryos for Fu or Ci have defects resembling Hh deficiency • Pka or Ptc mutants display ectopic expression of Hh target genes • CBP/p 300 is a co-activator for Ci-155

Role of microtuble association • Cos-2 tethers Ci and Fu to the microtubles • Cos-2 is a negative regulator of the pathway • Hh signaling leads to Cos-2 dissociation and phosphorylation of both Fu and Cos-2 • Unanswered questions: – How does Smo signal to the microtuble associated complex? – What exactly is in the complex?

Hedgehog signaling pathway From Munroe, et. al. (1999) Exp. Cell Res. 253: 25 -33

Vertebrate versions of Ci (the Gli proteins) • Behave differently in this pathway than Ci does in Drosophila • Gli-1, Gli-2 and Gli-3 appear to be transcriptionally regulated in response to Shh • See Munroe et. al. (1999) for several assorted speculations

Evidence for Sonic Hedgehog involvement in development • Mouse knockout results in abnormal limb development and cyclopia • Gli-3/Ci mutations lead to Grieg's cephalopolysyndactyly and other inherited diseases • Activating mutations can cause cancers (basal cell carcinomas-epidermal)

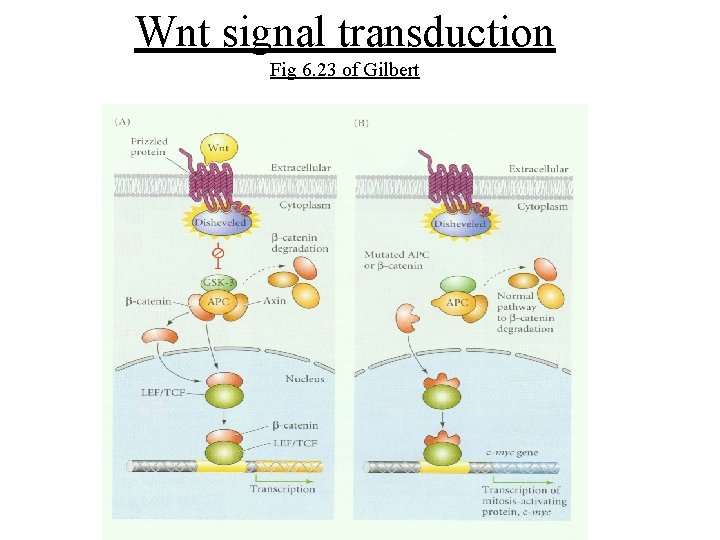
Wnt/wingless pathway • Ligands, Wnt-secreted glycoproteins – At least 16 members of the Wnt ligand family • Receptors are relatives of Frizzled (Frz) – 11 Frizzled homologs in vertebrates • Soluble Frz related proteins (Frps) exist – Antagonists of wnt signaling – Proposed to play a role in neural development

Wnt signal transduction Fig 6. 23 of Gilbert

Negative regulators of wnt signals • Glycogen synthase kinase (GSK)-3 b – Binds and phosphorylates several proteins in the wnt pathway to downregulate b-catenin • Adenomatous polyposis coli (APC) – Tumor suppressor in which mutations eliminate binding sites for Axin and b-catenin • Axin: Binds APC, b-catenin, GSK-3 b and dishevelled (Dvl) – Facilitates phosphorylation of APC and b-catenin by GSK-3 b

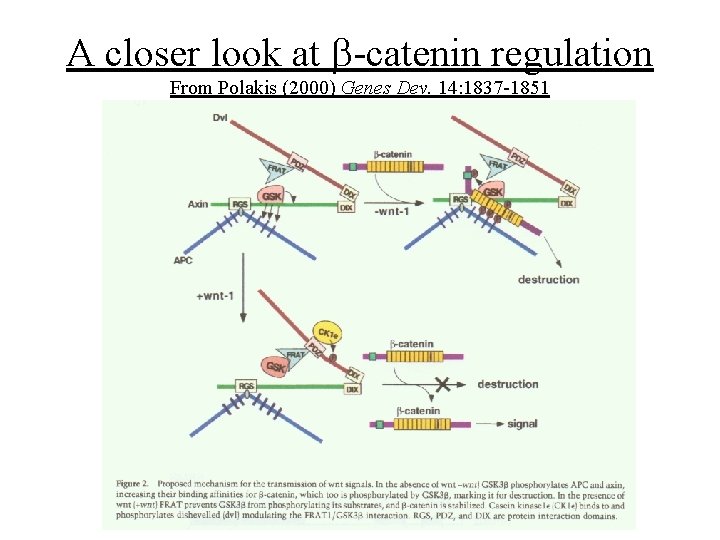
A closer look at b-catenin regulation From Polakis (2000) Genes Dev. 14: 1837 -1851

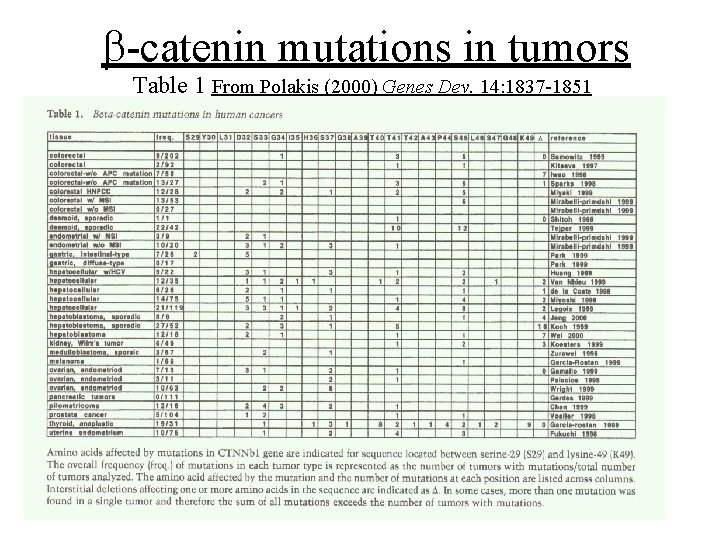
b-catenin mutations in tumors Table 1 From Polakis (2000) Genes Dev. 14: 1837 -1851

Clustered mutations in wnt signaling components affect negative interactions • b-catenin mutations in the N-terminal region affect amino acids necessary for its phosphorylation dependent interaction with protein degradation machinery • APC mutations affect axin and b-catenin binding • Axin mutations truncate the protein thus eliminating b-catenin binding sites

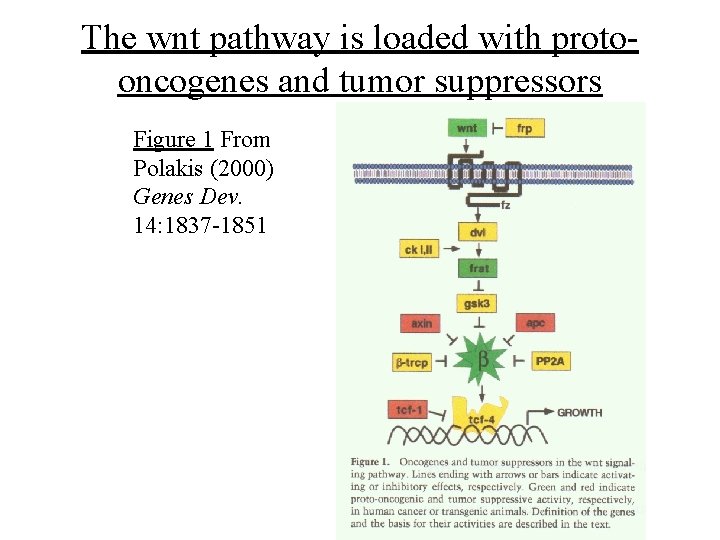
The wnt pathway is loaded with protooncogenes and tumor suppressors Figure 1 From Polakis (2000) Genes Dev. 14: 1837 -1851

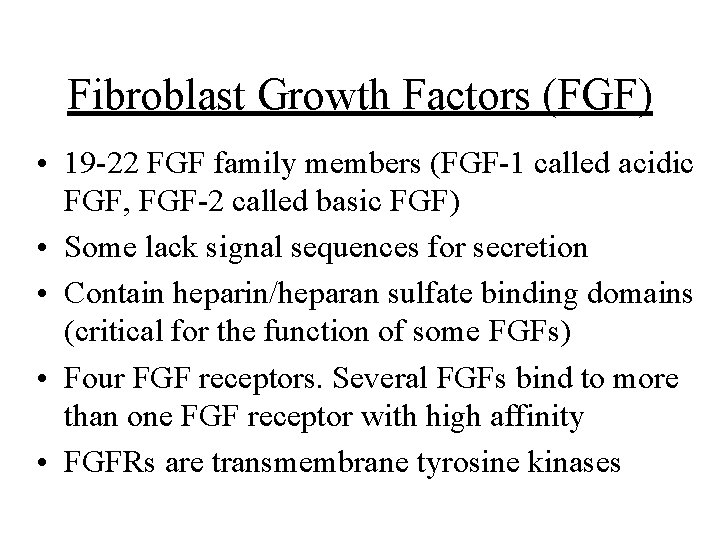
Fibroblast Growth Factors (FGF) • 19 -22 FGF family members (FGF-1 called acidic FGF, FGF-2 called basic FGF) • Some lack signal sequences for secretion • Contain heparin/heparan sulfate binding domains (critical for the function of some FGFs) • Four FGF receptors. Several FGFs bind to more than one FGF receptor with high affinity • FGFRs are transmembrane tyrosine kinases

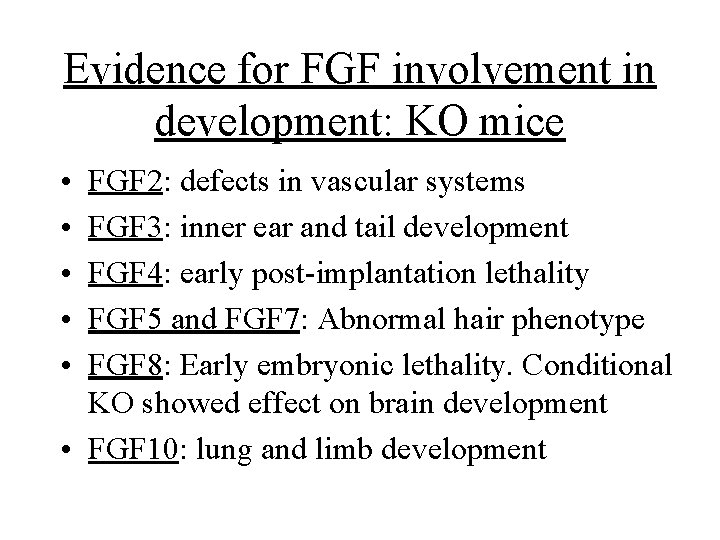
Evidence for FGF involvement in development: KO mice • • • FGF 2: defects in vascular systems FGF 3: inner ear and tail development FGF 4: early post-implantation lethality FGF 5 and FGF 7: Abnormal hair phenotype FGF 8: Early embryonic lethality. Conditional KO showed effect on brain development • FGF 10: lung and limb development

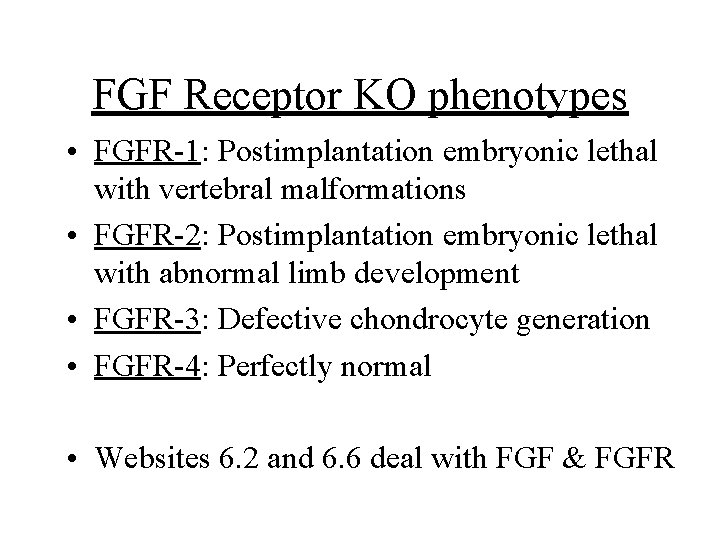
FGF Receptor KO phenotypes • FGFR-1: Postimplantation embryonic lethal with vertebral malformations • FGFR-2: Postimplantation embryonic lethal with abnormal limb development • FGFR-3: Defective chondrocyte generation • FGFR-4: Perfectly normal • Websites 6. 2 and 6. 6 deal with FGF & FGFR

Facts on Retinoic Acid • RA is a teratogen, causes birth defects • Interfering with RA biosynthesis causes developmental abnormalities linked to Hox gene expression • Exerts it effect through the retinoic acid receptors (RAR) which dimerizes with other steroid receptors to activate transcription • Important in development of limbs, neural tube and in anterior-posterior patterning