Heat shock Proteins HSPs n Heat shock proteins



- Slides: 12

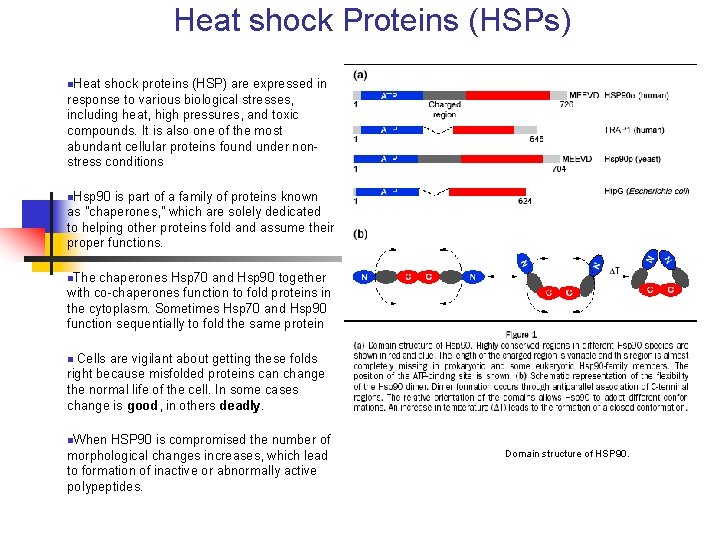
Heat shock Proteins (HSPs) n. Heat shock proteins (HSP) are expressed in response to various biological stresses, including heat, high pressures, and toxic compounds. It is also one of the most abundant cellular proteins found under nonstress conditions n. Hsp 90 is part of a family of proteins known as "chaperones, " which are solely dedicated to helping other proteins fold and assume their proper functions. n. The chaperones Hsp 70 and Hsp 90 together with co-chaperones function to fold proteins in the cytoplasm. Sometimes Hsp 70 and Hsp 90 function sequentially to fold the same protein n Cells are vigilant about getting these folds right because misfolded proteins can change the normal life of the cell. In some cases change is good, in others deadly. n. When HSP 90 is compromised the number of morphological changes increases, which lead to formation of inactive or abnormally active polypeptides. Domain structure of HSP 90.

Mad Cows, People & yeast n n n What do "mad cows, " people with neurodegenerative diseases and an unusual type of yeast have in common? They are all experiencing the effects of misfolded proteins, according to HHMI investigator Susan Lindquist of the University of Chicago. Her research identified a role for HSP 90 in the process of evolution. They have reported that fruit flies that make too little of the Hsp 90 protein develop dramatic deformities, such as an extra antenna, additional bristles, notched wings or malformed eyes. The defects result from multiple hidden variations in the genome. When affected flies are interbred, these factors are enriched and subsequent generations have the same deformities, even though they have normal levels of Hsp 90. Source: http: //www. hhmi. org/annual 98/research/madcow. html Susan Lindquist University of Chicago "My view is that molecular chaperones are a way of changing the traits of an organism that arose very early in evolution. They might be as old as RNA and DNA. "

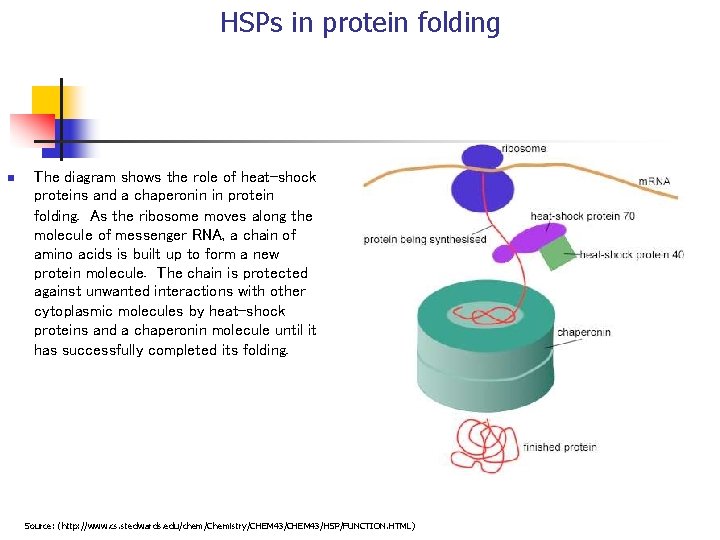
HSPs in protein folding n The diagram shows the role of heat-shock proteins and a chaperonin in protein folding. As the ribosome moves along the molecule of messenger RNA, a chain of amino acids is built up to form a new protein molecule. The chain is protected against unwanted interactions with other cytoplasmic molecules by heat-shock proteins and a chaperonin molecule until it has successfully completed its folding. Source: (http: //www. cs. stedwards. edu/chem/Chemistry/CHEM 43/HSP/FUNCTION. HTML)

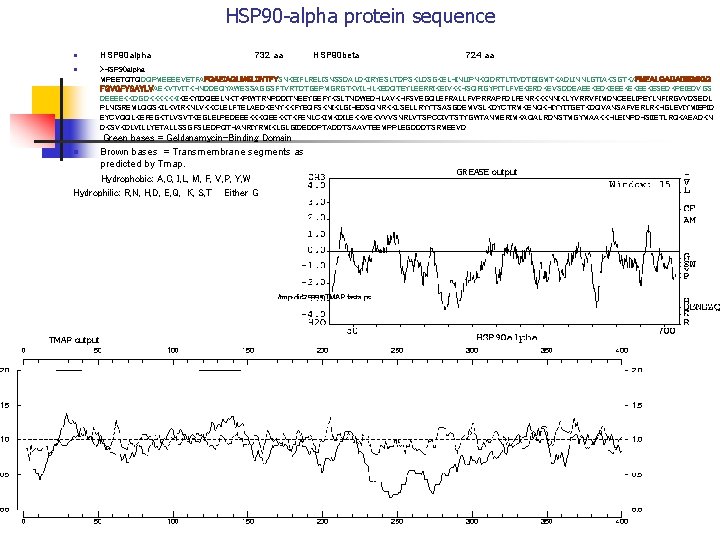
HSP 90 -alpha protein sequence n n HSP 90 alpha 732 aa HSP 90 beta 724 aa >HSP 90 alpha MPEETQTQDQPMEEEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYESLTDPSKLDSGKELHINLIPNKQDRTLTIVDTGIGMTKADLINNLGTIAKSGTKAFMEALQAGADISMIGQ FGVGFYSAYLVAEKVTVITKHNDDEQYAWESSAGGSFTVRTDTGEPMGRGTKVILHLKEDQTEYLEERRIKEIVKKHSQFIGYPITLFVEKERDKEVSDDEAEEKEDKEEEKEKEEKESEDKPEIEDVGS DEEEEKKDGDKKKKKKIKEKYIDQEELNKTKPIWTRNPDDITNEEYGEFYKSLTNDWEDHLAVKHFSVEGQLEFRALLFVPRRAPFDLFENRKKKNNIKLYVRRVFIMDNCEELIPEYLNFIRGVVDSEDL PLNISREMLQQSKILKVIRKNLVKKCLELFTELAEDKENYKKFYEQFSKNIKLGIHEDSQNRKKLSELLRYYTSASGDEMVSLKDYCTRMKENQKHIYYITGETKDQVANSAFVERLRKHGLEVIYMIEPID EYCVQQLKEFEGKTLVSVTKEGLELPEDEEEKKKQEEKKTKFENLCKIMKDILEKKVEKVVVSNRLVTSPCCIVTSTYGWTANMERIMKAQALRDNSTMGYMAAKKHLEINPDHSIIETLRQKAEADKN DKSVKDLVILLYETALLSSGFSLEDPQTHANRIYRMIKLGLGIDEDDPTADDTSAAVTEEMPPLEGDDDTSRMEEVD Green bases = Geldanamycin-Binding Domain Brown bases = Transmembrane segments as predicted by Tmap. Hydrophobic: A, C, I, L, M, F, V, P, Y, W Hydrophilic: R, N, H, D, E, Q, K, S, T Either G n TMAP output GREASE output

BLASTp n n n With the HSP 90 sequence in hand we used Blastp to find homologous sequences We were surprised to find a lot of homologous sequences across many species like Humans, Chicken, Pig, Mouse, Horse, Fish, Coral, fruit fly, mosquito, nematode, & even crops like rice, maize & tobacco. The first 100 matches had e-values ranging from 0 to e-153, so they were *very* strong matches indicating a high degree of conservation of the protein through evolution. ID Name 304882 heat shock 90 k. Da protein 1, alpha [Homo sapiens] N. . . Score Evalue 1247 0. 0 352285 heat shock protein 1, alpha [Mus musculus] NP_0346. . . 825 0. 0 761972 heat shock protein 86 [Rattus norvegicus] NP_78693. . . 825 0. 0 341493 heat shock protein 90 A [Cricetulus griseus] AAA 369. . . 817 0. 0 609431 heat shock protein 90 - chicken 816 0. 0 609432 heat shock protein 84 - mouse 745 0. 0 449511 (Q 9 W 6 K 6) Heat shock protein hsp 90 beta [Salmo sala. . . 731 0. 0 459017 heat shock protein hsp 90 [Oncorhynchus tshawytscha. . . 730 0. 0 446434 heat shock protein hsp 90 beta [Danio rerio] AAC 2156. . . 729 0. 0 361999 heat shock protein 90 [Rattus sp. ] AAB 23369. 1 [S 45. . . 724 0. 0 460597 heat shock protein 90 [Pleurodeles waltl] AAA 92343. . . 719 0. 0 738604 90 -k. Da heat shock protein [Bombyx mori] BAB 41209. 1. . . 712 0. 0 146263 Heat shock protein 83 CG 1242 -PA [Drosophila melano. . . 669 0. 0 755572 heat shock protein 90 [Dendronephthya klunzingeri]. . . 662 0. 0 226533 (P 33126) Heat shock protein 82 [Oryza sativa (Rice)] 612 e-174 1888761 heat shock protein 82 - common tobacco (fragment) 612 e-174 252633 heat shock protein [Arabidopsis thaliana] CAA 72513. . . 600 e-170 236351 (Q 9 XGF 1) HSP 80 -2 [Triticum aestivum (Wheat)] 598 e-169 283559 (Q 08277) Heat shock protein 82 [Zea mays (Maize)] 593 e-168 152674 heat shock protein 86 [Plasmodium falciparum] AAA 6. . . 591 e-167 1899880 (Q 8 LLI 6) Heat shock protein Hsp 90 [Achlya ambisex. . . 579 e-164 245912 heat shock protein 90 [Lycopersicon esculentum] AA. . . 544 e-153

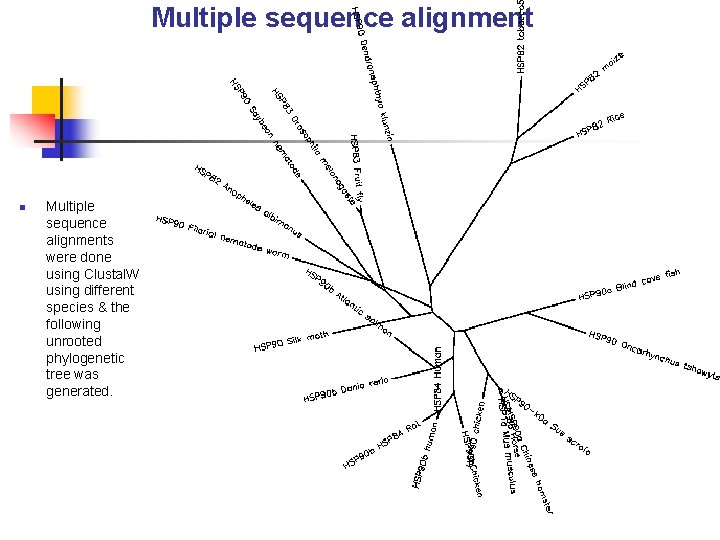
Multiple sequence alignment n Multiple sequence alignments were done using Clustal. W using different species & the following unrooted phylogenetic tree was generated.

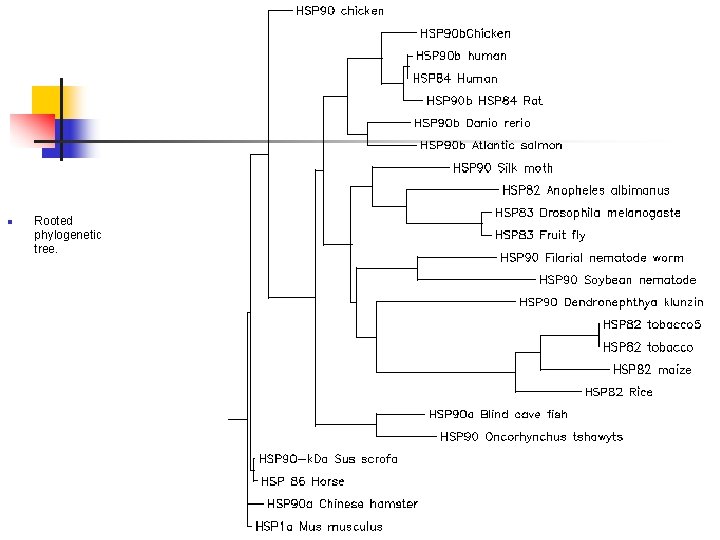
n Rooted phylogenetic tree.

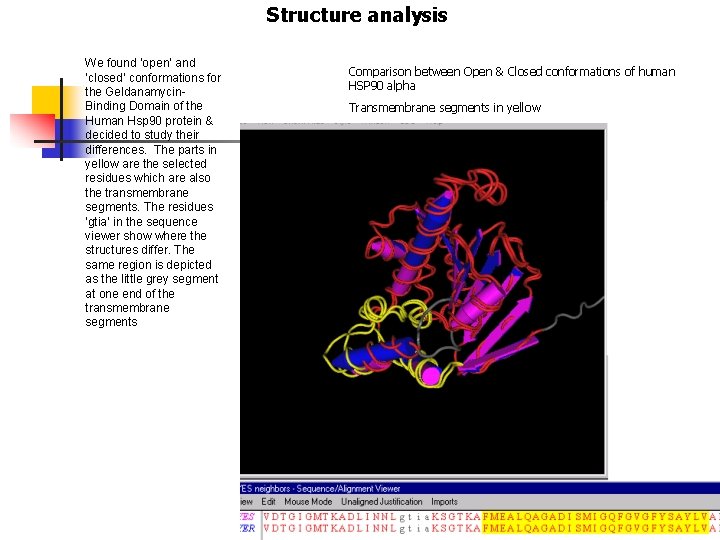
Structure analysis We found ‘open’ and ‘closed’ conformations for the Geldanamycin. Binding Domain of the Human Hsp 90 protein & decided to study their differences. The parts in yellow are the selected residues which are also the transmembrane segments. The residues ‘gtia’ in the sequence viewer show where the structures differ. The same region is depicted as the little grey segment at one end of the transmembrane segments Comparison between Open & Closed conformations of human HSP 90 alpha Transmembrane segments in yellow

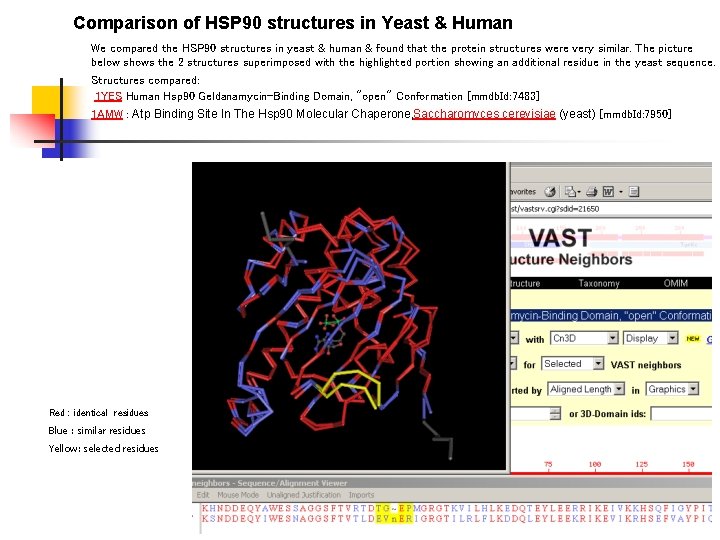
Comparison of HSP 90 structures in Yeast & Human We compared the HSP 90 structures in yeast & human & found that the protein structures were very similar. The picture below shows the 2 structures superimposed with the highlighted portion showing an additional residue in the yeast sequence. Structures compared: 1 YES Human Hsp 90 Geldanamycin-Binding Domain, "open" Conformation [mmdb. Id: 7483] 1 AMW : Atp Binding Site In The Hsp 90 Molecular Chaperone, Saccharomyces cerevisiae (yeast) [mmdb. Id: 7950] Red : identical residues Blue : similar residues Yellow: selected residues

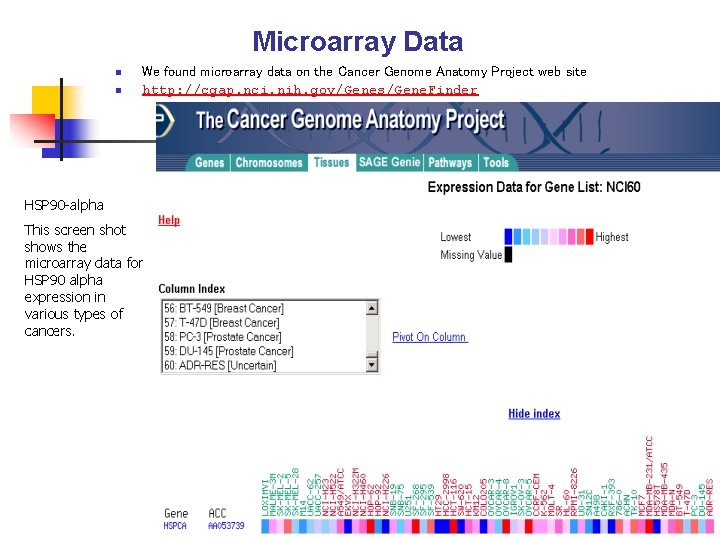
Microarray Data n n We found microarray data on the Cancer Genome Anatomy Project web site http: //cgap. nci. nih. gov/Genes/Gene. Finder HSP 90 -alpha This screen shot shows the microarray data for HSP 90 alpha expression in various types of cancers.

HSP 90 -beta This screen shot shows the microarray data for HSP 90 beta expression in various types of cancers.

Current Studies on HSP 90 Changes in protein conformation are involved in some of the most devastating and intractable diseases. “Studies in yeast may help us decipher the fundamental nature of these disorders, including Creutzfeldt-Jakob, Alzheimer’s, Huntington’s, and Parkinson’s disease in humans and mad cow disease in cattle. Several of the protein culprits are being imported into yeast, which allows for the manipulation & study of their folding transitions & testing of therapeutic strategies. (http: //www. wi. mit. edu/nap/pdfs/Directors_Report/dir_lindquist 02. pdf) Conclusion: HSP 90 is a powerful evolutionary mechanism that ensures apparent genetic stability at physiological conditions and at the same time allows the mutations that could rapidly become manifest under stress. References: http: //www. stanford. edu/class/gene 211/hsp 90_search http: //www. chemie. tu-muenchen. de/biotech/en/hsp 90 -e. html http: //www. hhmi. org/annual 98/research/madcow. html www. ashland. edu/~kstine/Research/Stress%20 proteins. pdf
 Hangman fracture
Hangman fracture Subacute combined degeneration
Subacute combined degeneration Diferencia entre shock medular y shock neurogenico
Diferencia entre shock medular y shock neurogenico Spinal shock vs neurogenic shock
Spinal shock vs neurogenic shock Spinal shock vs neurogenic shock
Spinal shock vs neurogenic shock Heat shock response
Heat shock response Protein monomers
Protein monomers Biomedical importance of proteins
Biomedical importance of proteins Structural role of proteins
Structural role of proteins Peripheral proteins
Peripheral proteins Globular vs fibrous proteins
Globular vs fibrous proteins Carbohydrates in milk
Carbohydrates in milk Precipitation of proteins by strong mineral acids
Precipitation of proteins by strong mineral acids