Gwyndaf Evans 1 Graeme Winter 1 David Waterman
- Slides: 1

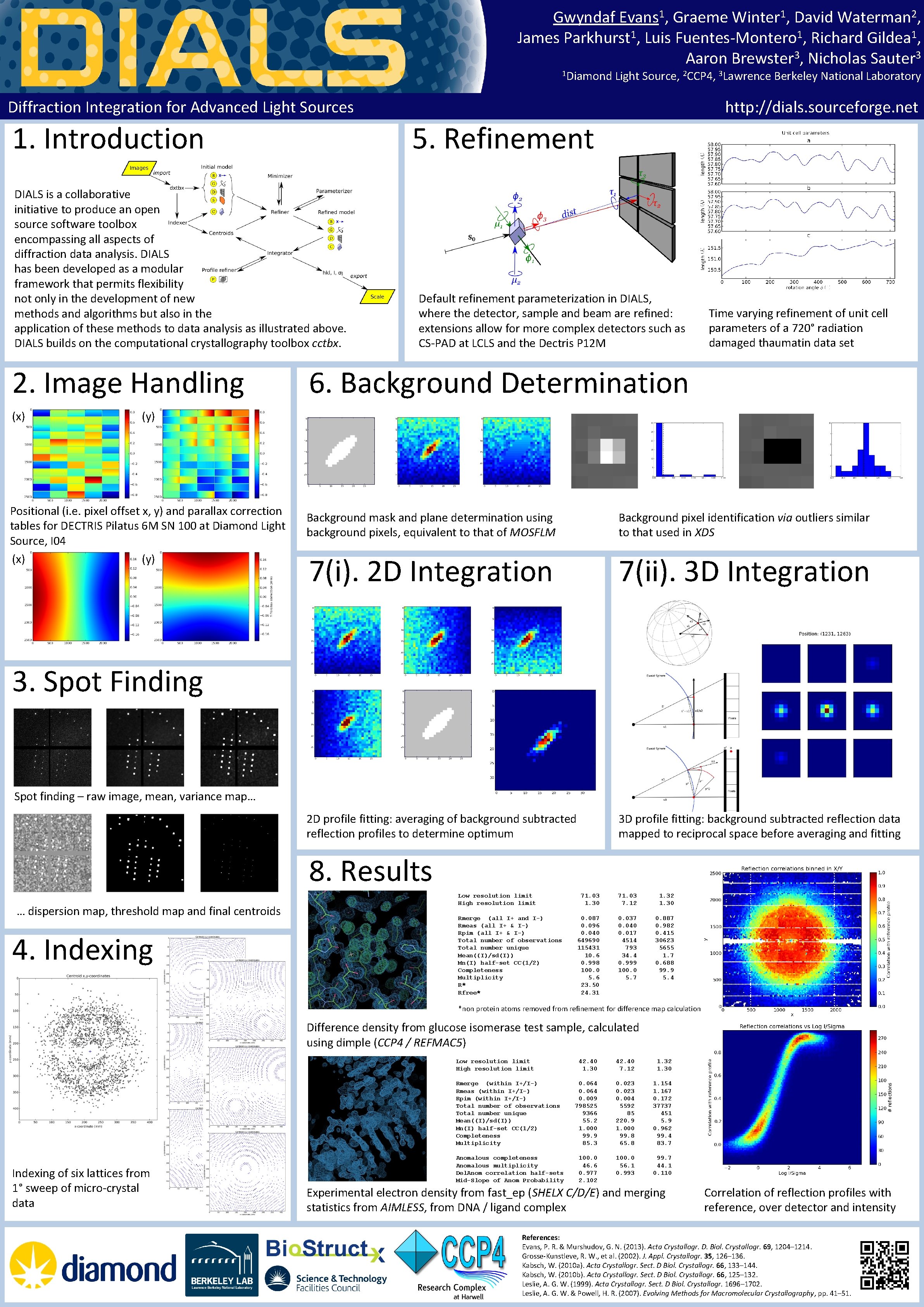
Gwyndaf Evans 1, Graeme Winter 1, David Waterman 2, James Parkhurst 1, Luis Fuentes-Montero 1, Richard Gildea 1, Aaron Brewster 3, Nicholas Sauter 3, 1 Diamond Light Source, 2 CCP 4, 3 Lawrence Berkeley National Laboratory Diffraction Integration for Advanced Light Sources 1. Introduction 5. Refinement DIALS is a collaborative initiative to produce an open source software toolbox encompassing all aspects of diffraction data analysis. DIALS has been developed as a modular framework that permits flexibility not only in the development of new methods and algorithms but also in the application of these methods to data analysis as illustrated above. DIALS builds on the computational crystallography toolbox cctbx. 2. Image Handling (x) http: //dials. sourceforge. net Default refinement parameterization in DIALS, where the detector, sample and beam are refined: extensions allow for more complex detectors such as CS-PAD at LCLS and the Dectris P 12 M Time varying refinement of unit cell parameters of a 720° radiation damaged thaumatin data set 6. Background Determination (y) Positional (i. e. pixel offset x, y) and parallax correction tables for DECTRIS Pilatus 6 M SN 100 at Diamond Light Source, I 04 (x) (y) Background mask and plane determination using background pixels, equivalent to that of MOSFLM Background pixel identification via outliers similar to that used in XDS 7(i). 2 D Integration 7(ii). 3 D Integration 2 D profile fitting: averaging of background subtracted reflection profiles to determine optimum 3 D profile fitting: background subtracted reflection data mapped to reciprocal space before averaging and fitting 3. Spot Finding Spot finding – raw image, mean, variance map… 8. Results … dispersion map, threshold map and final centroids 4. Indexing Low resolution limit High resolution limit Rmerge (all I+ and I-) Rmeas (all I+ & I-) Rpim (all I+ & I-) Total number of observations Total number unique Mean((I)/sd(I)) Mn(I) half-set CC(1/2) Completeness Multiplicity R* Rfree* 71. 03 1. 30 71. 03 7. 12 1. 30 0. 087 0. 096 0. 040 649690 115431 10. 6 0. 998 100. 0 5. 6 23. 50 24. 31 0. 037 0. 040 0. 017 4514 793 34. 4 0. 999 100. 0 5. 7 0. 887 0. 982 0. 415 30623 5655 1. 7 0. 688 99. 9 5. 4 *non protein atoms removed from refinement for difference map calculation Difference density from glucose isomerase test sample, calculated using dimple (CCP 4 / REFMAC 5) Low resolution limit High resolution limit Rmerge (within I+/I-) Rmeas (within I+/I-) Rpim (within I+/I-) Total number of observations Total number unique Mean((I)/sd(I)) Mn(I) half-set CC(1/2) Completeness Multiplicity Indexing of six lattices from 1° sweep of micro-crystal data Anomalous completeness Anomalous multiplicity Del. Anom correlation half-sets Mid-Slope of Anom Probability 42. 40 1. 30 42. 40 7. 12 1. 30 0. 064 0. 009 798525 9366 55. 2 1. 000 99. 9 85. 3 0. 023 0. 004 5592 85 220. 9 1. 000 99. 8 65. 8 1. 154 1. 167 0. 172 37737 451 5. 9 0. 962 99. 4 83. 7 100. 0 46. 6 0. 977 2. 102 100. 0 56. 1 0. 993 99. 7 44. 1 0. 110 Experimental electron density from fast_ep (SHELX C/D/E) and merging statistics from AIMLESS, from DNA / ligand complex Correlation of reflection profiles with reference, over detector and intensity References: Evans, P. R. & Murshudov, G. N. (2013). Acta Crystallogr. D. Biol. Crystallogr. 69, 1204– 1214. Grosse-Kunstleve, R. W. , et al. (2002). J. Appl. Crystallogr. 35, 126– 136. Kabsch, W. (2010 a). Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 133– 144. Kabsch, W. (2010 b). Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 125– 132. Leslie, A. G. W. (1999). Acta Crystallogr. Sect. D Biol. Crystallogr. 1696– 1702. Leslie, A. G. W. & Powell, H. R. (2007). Evolving Methods for Macromolecular Crystallography, pp. 41– 51.