GSEAPro Tutorial Gene Set Enrichment Analysis for Prokaryotes






- Slides: 9

GSEA-Pro Tutorial Gene Set Enrichment Analysis for Prokaryotes Anne de Jong, University of Groningen

Introduction � The main principle of a Gene Set Enrichment Analysis (GSEA) is to discover which biological function is or functions are overrepresented in a set of genes or proteins resulting from an -omics (e. g. RNA-Seq) analysis. � GSEA-Pro uses the Genome 2 D database that describes the relation between genes/proteins and functions (functional classification) of all Ref. Seq and Genbank complete genomes (>20 k genomes). � GSEA-Pro uses multiple classification; COG, GO, KEGG, PFAM, Inter. Pro, Superfamily and Keywords. � GSEA-Pro only allows locus-tags as ID for genes as well as for proteins

Introduction � Overview of Functional Analysis of Genes Sets Transcriptomics Proteomics Metagenomics -omics One or multiple sets of Genes Unravel the biological function of a “Gene Set”

Input � STEP 1: Select Genome � GSEA-Pro offers the choice between Ref. Seq or Genbank. Be aware that for the same strain the locus-tags might differ between Ref. Seq and Genbank, commonly the Ref. Seq locus-tags contain the 2 -letter code 'RS'. Conversion between Ref. Seq and Genbank locus-tags is supported at http: //genome 2 d. molgenrug. nl/ � STEP 2: Four types of data tables can be used as input � Single list of locus-tags: This is a bare list of 'Top Hits' genes (as locus-tags) deduced from transcriptome or proteome analysis results. Meaning only those that are significant are posted here � Single list of locus-tags with ratio values: The first column contains the locus-tags, the second ratio values generated by differential expression (DE) analysis. In this case the cutoff values are used or auto detected by GSEA-Pro. � Experiments: GSEA-Pro can handle multiple experiments (e. g. derived from time series). Also here the cutoff values can be set or auto detected. � Clustering: Clustering algorithms will group genes showing similar behavior over perturbation experiments or time series. GSEA-Pro will handle each cluster as a gene set and will show the biological function of each cluster. The first column of the input table should contain the locustags and the column with cluster ID's should have the obligatory header “cluster. ID”.

Input � Step 3: Examples of input data tables Tables can be uploaded to the webserver as tab delimited file or by copy and paste directly from e. g. Excel Single list + ratio data Experiments Clustering [ value columns will be ignored ]

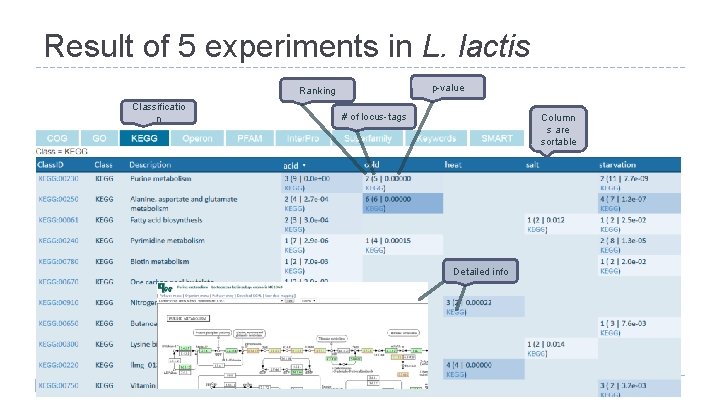
Result of 5 experiments in L. lactis p-value Ranking Classificatio n # of locus-tags Column s are sortable Detailed info


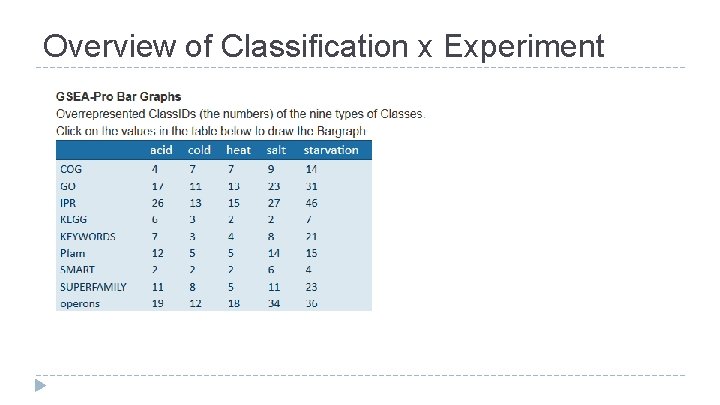
Overview of Classification x Experiment
