Group Testing and New Algorithmic Applications Ely Porat

![More Motivations • • • Syphilis, HIV [Dor 43] Mapping genomes [BLC 91, BBK+95, More Motivations • • • Syphilis, HIV [Dor 43] Mapping genomes [BLC 91, BBK+95,](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-6.jpg)





![Group testing for Pattern Matching • Pattern matching with wildcards – O(nlogm) [CH 02] Group testing for Pattern Matching • Pattern matching with wildcards – O(nlogm) [CH 02]](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-21.jpg)




![Reducing sketching space [BP 10] Instead of Additional pairwise independent hash It was discover Reducing sketching space [BP 10] Instead of Additional pairwise independent hash It was discover](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-38.jpg)
![Reducing sketching space [BP 10] Our algorithm estimates Reducing sketching space [BP 10] Our algorithm estimates](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-39.jpg)
![Reducing sketching space even farther [BP 10] We usually interesting in the case that Reducing sketching space even farther [BP 10] We usually interesting in the case that](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-40.jpg)
![Reducing sketching space even farther [BP 10] We usually interesting in the case that Reducing sketching space even farther [BP 10] We usually interesting in the case that](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-41.jpg)
![Removing the min wise independent requirement [BP 11] • [KNW 10] gave bits sketch Removing the min wise independent requirement [BP 11] • [KNW 10] gave bits sketch](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-42.jpg)
![Removing the min wise independent requirement [BP 11] Using F 2 instead of F Removing the min wise independent requirement [BP 11] Using F 2 instead of F](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-43.jpg)



![Selective verification [PW 12] Packeti S'i If we have both signatures we can choose Selective verification [PW 12] Packeti S'i If we have both signatures we can choose](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-55.jpg)
![Analysis 1 n+m 2 dn Pr[one block pass the test]<qn/qdn=q-(d-1)n Pr[r/2 out of r Analysis 1 n+m 2 dn Pr[one block pass the test]<qn/qdn=q-(d-1)n Pr[r/2 out of r](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-59.jpg)
![Analysis 1 n+m 2 dn Pr[one block pass the test]<qn/qdn=q-(d-1)n Pr[r/2 out of r Analysis 1 n+m 2 dn Pr[one block pass the test]<qn/qdn=q-(d-1)n Pr[r/2 out of r](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-60.jpg)

- Slides: 61

Group Testing and New Algorithmic Applications Ely Porat Bar-Ilan University

Theory of Big data Coding theory Pattern matching Group testing Compressive sensing Game theory Distributed

Theory of Big data Succinct data structures Sketching & LSH Bloom filters Big Databases Streaming algorithm

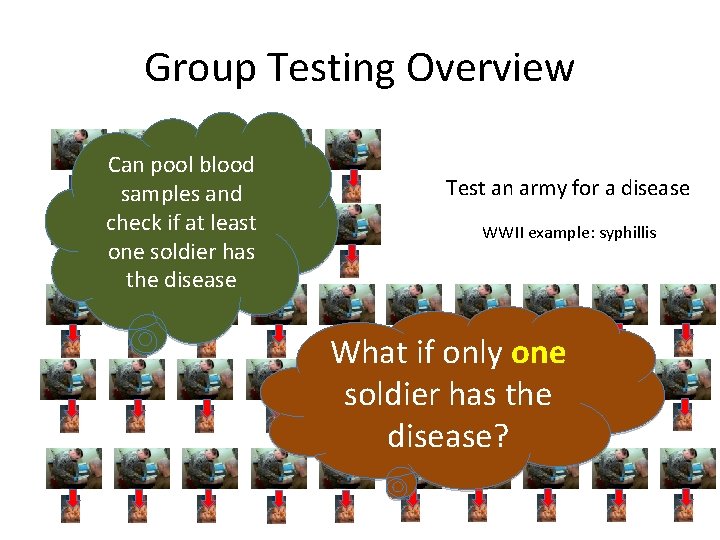
Group Testing Overview Test soldier for a disease WWII example: syphillis

Group Testing Overview Can pool blood samples and check if at least one soldier has the disease Test an army for a disease WWII example: syphillis What if only one soldier has the disease?
![More Motivations Syphilis HIV Dor 43 Mapping genomes BLC 91 BBK95 More Motivations • • • Syphilis, HIV [Dor 43] Mapping genomes [BLC 91, BBK+95,](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-6.jpg)
More Motivations • • • Syphilis, HIV [Dor 43] Mapping genomes [BLC 91, BBK+95, TJP 00] Quality control in product testing [SG 59] Searching files in storage systems [KS 64] Sequential screening of experimental variables [Li 62] Efficient contention resolution algorithms for multiple access communication [KS 64, Wol 85] Data compression [HL 00] Software testing [BG 02, CDFP 97] DNA sequencing [PL 94] Molecular biology [DH 00, FKKM 97, ND 00, BBKT 96]

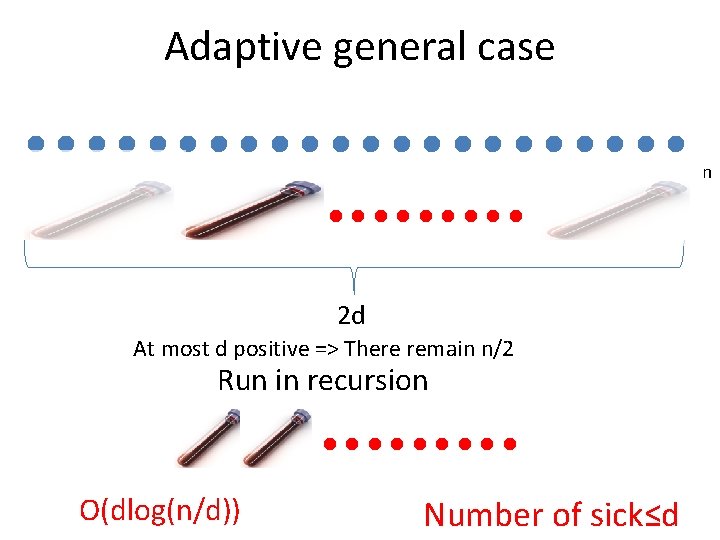
Adaptive group testing Number of sick d≤ 2

Adaptive general case n 2 d At most d positive => There remain n/2 Run in recursion O(dlog(n/d)) Number of sick≤d

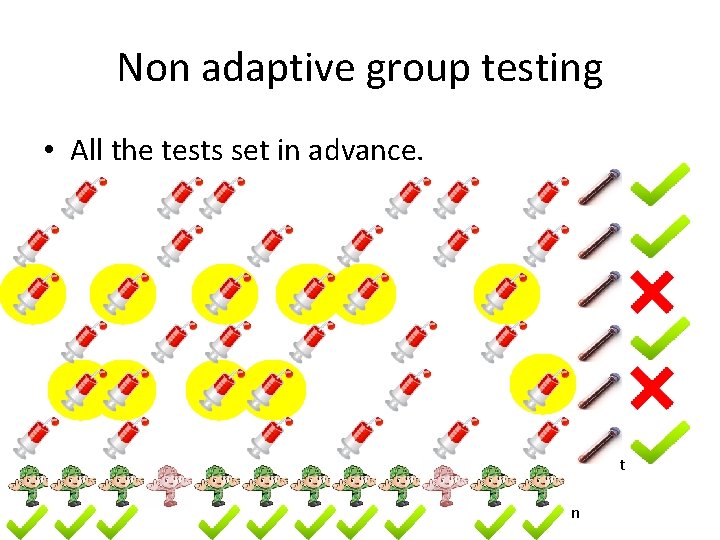
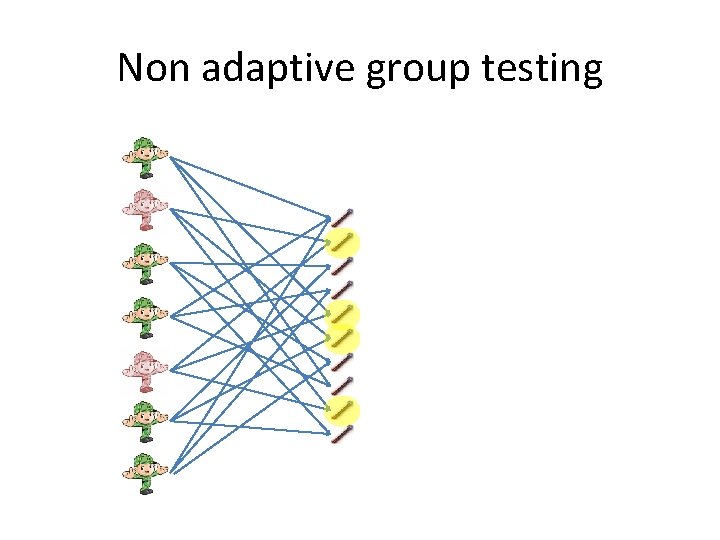
Non adaptive group testing • All the tests set in advance. t n

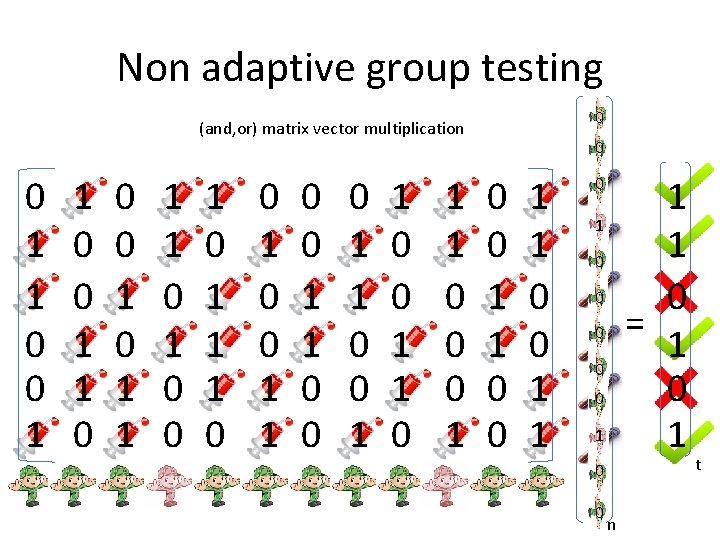
Non adaptive group testing 0 (and, or) matrix vector multiplication 0 1 1 0 0 0 1 1 1 1 0 0 1 1 0 0 0 1 1 0 0 1 1 1 1 0 = 1 0 1 0 0 n t

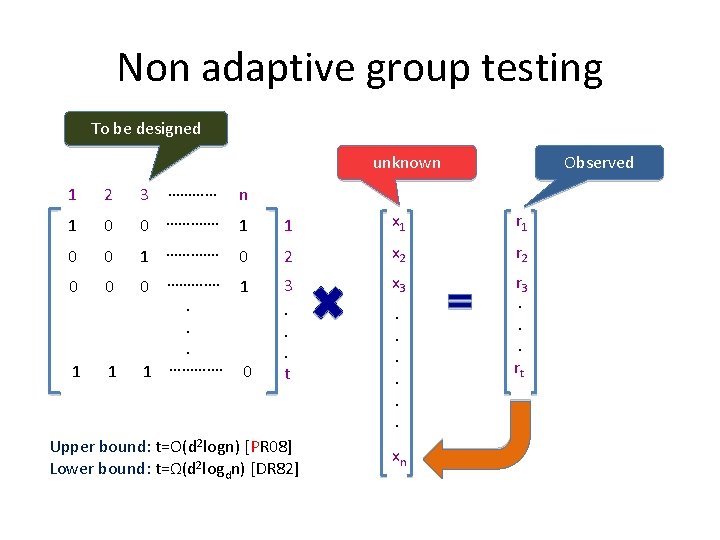
Non adaptive group testing To be designed unknown Observed 1 2 3 ………… n 1 0 1 1 x 1 r 1 0 0 0 …………. 1 …………. 0 2 x 2 r 2 0 0 0 …………. 1. . . 1 …………. 0 3. . . t x 3 r 3. . . rt 1 1 Upper bound: t=O(d 2 logn) [PR 08] Lower bound: t=Ω(d 2 logdn) [DR 82] . . . xn

Non adaptive group testing

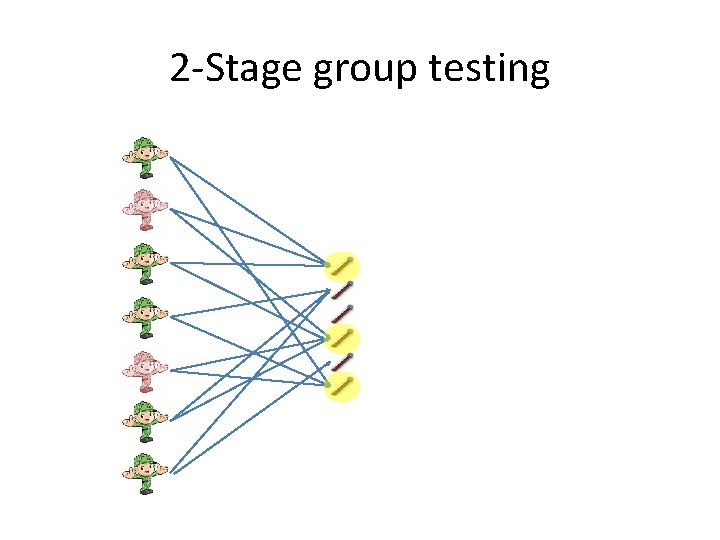
2 -Stage group testing

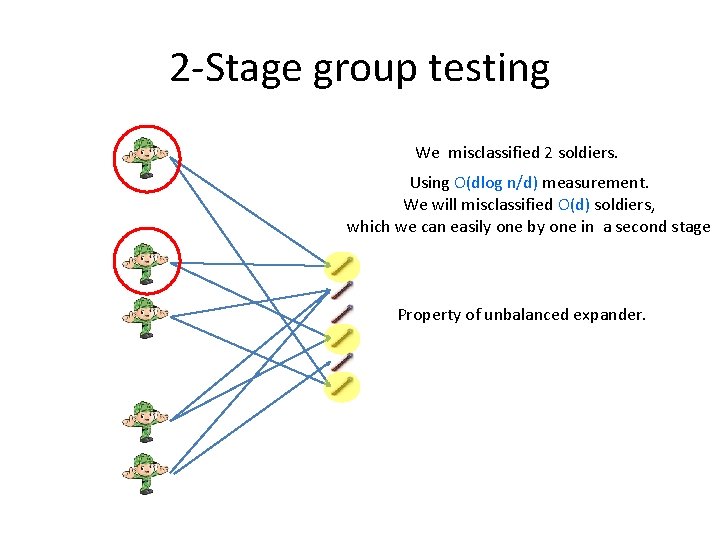
2 -Stage group testing We misclassified 2 soldiers. Using O(dlog n/d) measurement. We will misclassified O(d) soldiers, which we can easily one by one in a second stage Property of unbalanced expander.

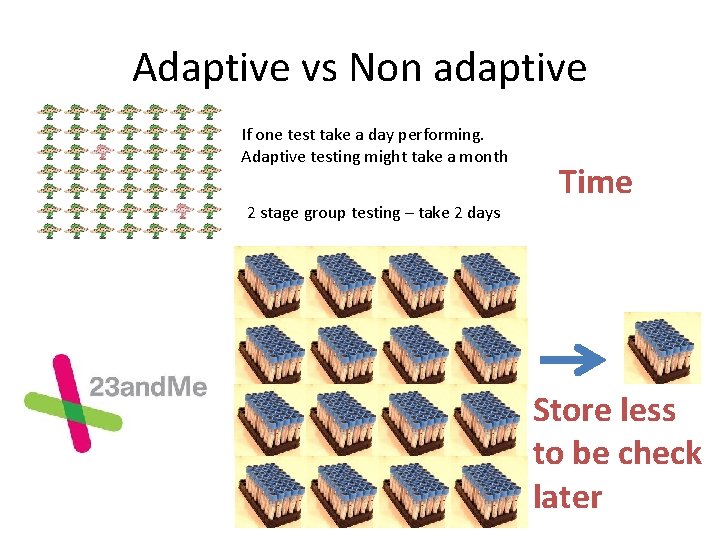
Adaptive vs Non adaptive If one test take a day performing. Adaptive testing might take a month 2 stage group testing – take 2 days Time Store less to be check later

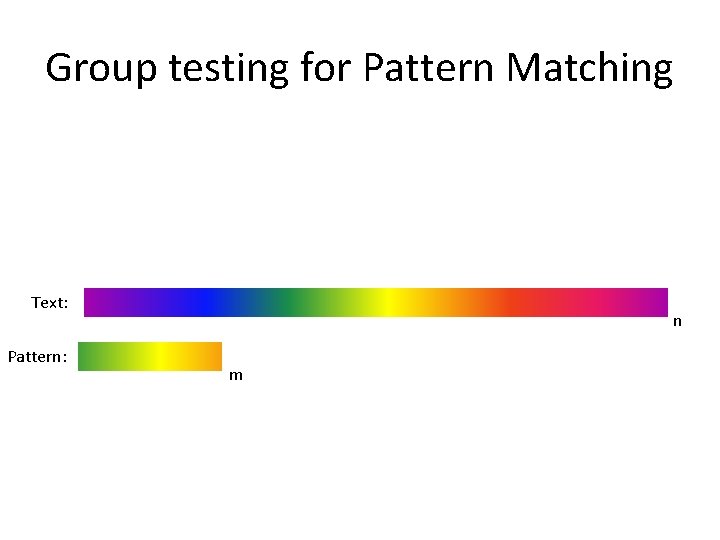
Group testing for Pattern Matching Text: Pattern: n m

Group testing for Pattern Matching Part of 20 M€ consortium project which is supported by MOI (cyber security)

Motivation… • Stock market

Motivation. . • Espionage The rest we monitor

Motivation… • Viruses and malware Software solutions: Snort: 73. 5 Mb Clam. AV: 1. 48 Gb Using TCAMs: Snort: 680 Kb Clam. AV: 25 Mb Our solution (software): Snort: 51 Kb Clam. AV: 216 Kb
![Group testing for Pattern Matching Pattern matching with wildcards Onlogm CH 02 Group testing for Pattern Matching • Pattern matching with wildcards – O(nlogm) [CH 02]](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-21.jpg)
Group testing for Pattern Matching • Pattern matching with wildcards – O(nlogm) [CH 02] • Up to k mismatches [CEPR 07, CEPR 09]. Text: n Pattern: m • Sketching hamming distance [PL 07, AGGP 13]. • Pattern matching in the streaming model [PP 09]

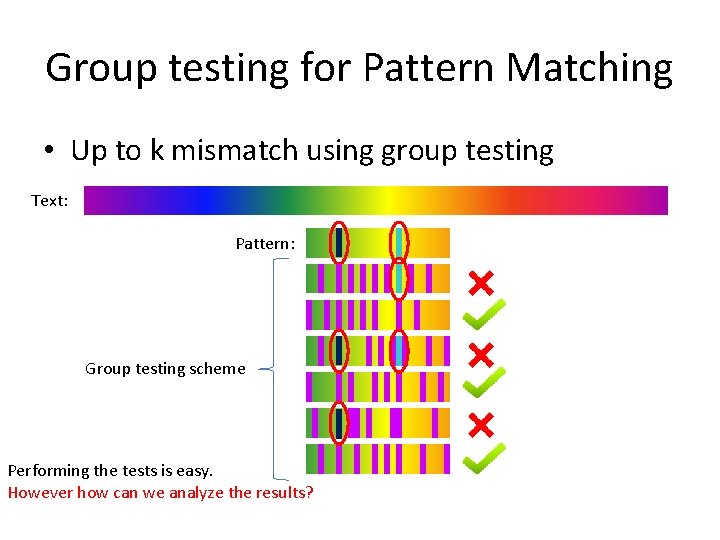
Group testing for Pattern Matching • Up to k mismatch using group testing Text: Pattern: Group testing scheme Performing the tests is easy. However how can we analyze the results?

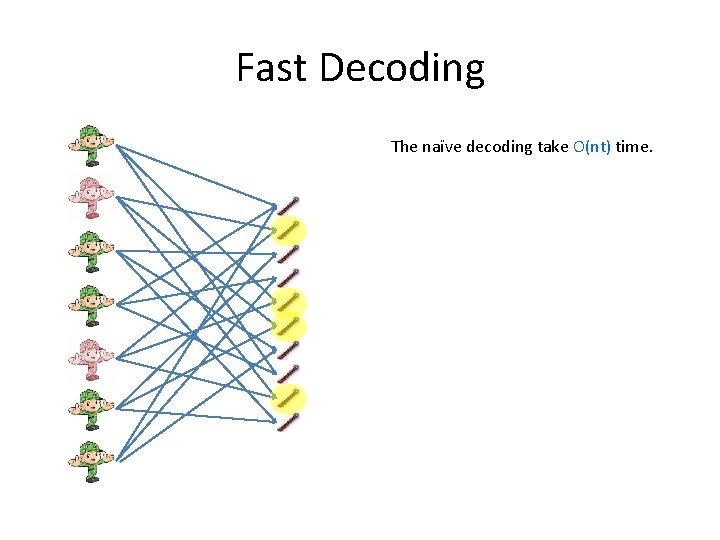
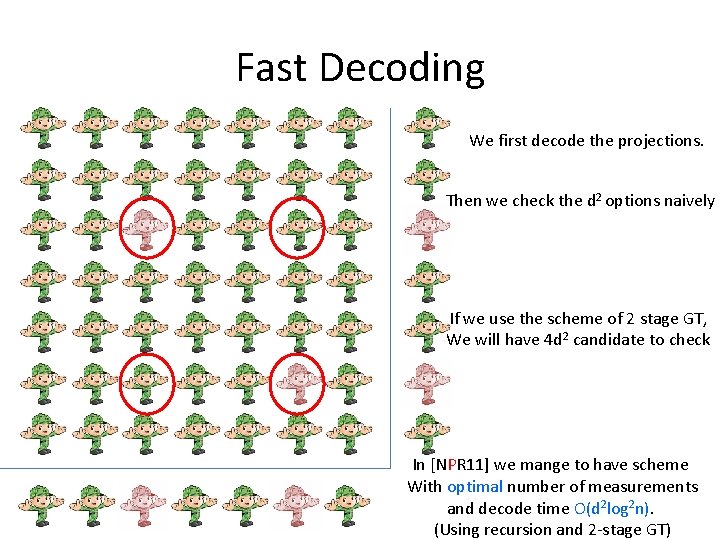
Fast Decoding The naïve decoding take O(nt) time.

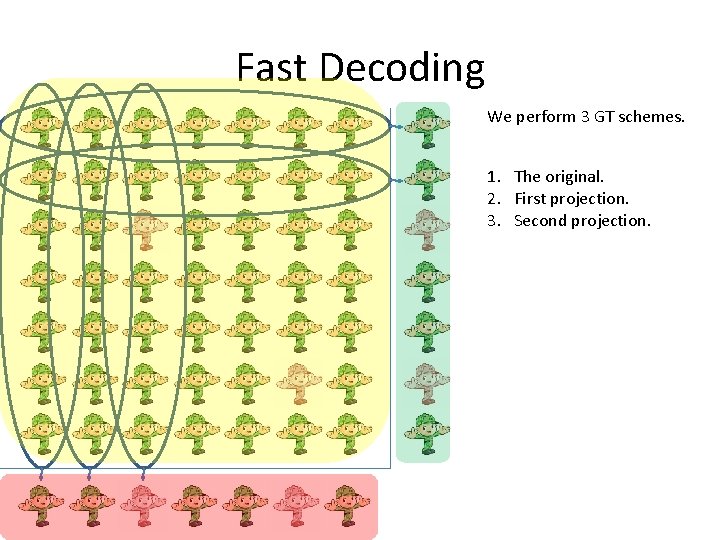
Fast Decoding We perform 3 GT schemes. 1. The original. 2. First projection. 3. Second projection.

Fast Decoding We first decode the projections. Then we check the d 2 options naively If we use the scheme of 2 stage GT, We will have 4 d 2 candidate to check In [NPR 11] we mange to have scheme With optimal number of measurements and decode time O(d 2 log 2 n). (Using recursion and 2 -stage GT)

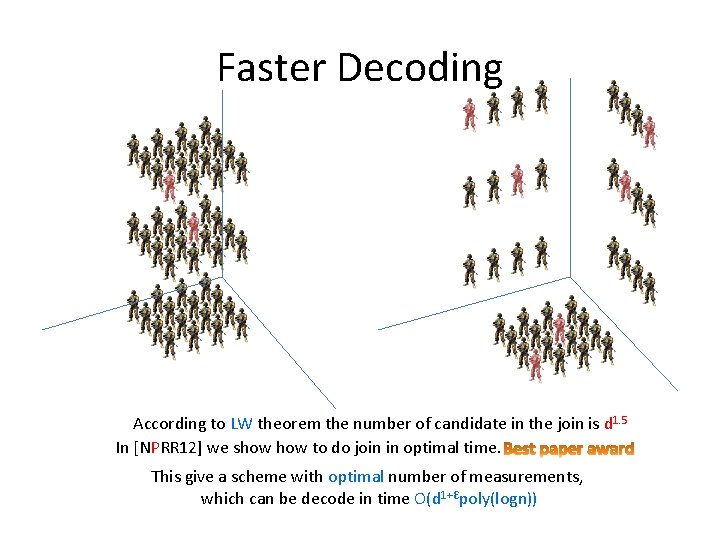
Faster Decoding According to LW theorem the number of candidate in the join is d 1. 5 In [NPRR 12] we show to do join in optimal time. This give a scheme with optimal number of measurements, which can be decode in time O(d 1+Ԑpoly(logn))

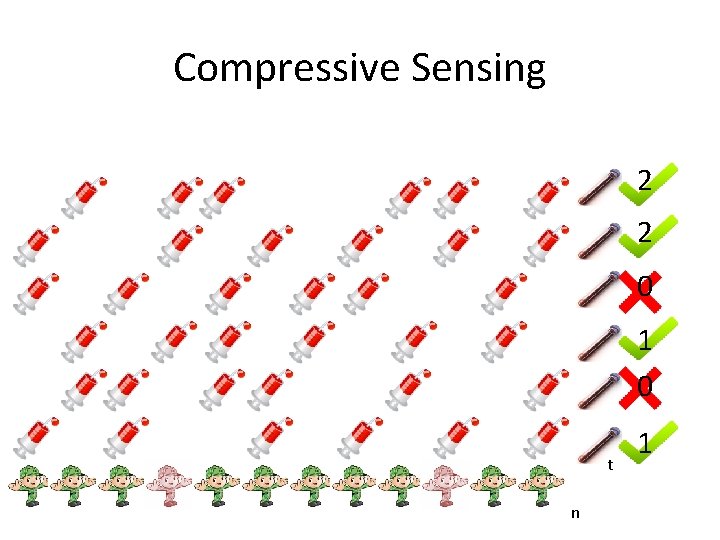
Compressive Sensing 2 2 0 1 0 t n 1

Compressive Sensing 0 0 0 1 1 0 0 0 1 1 1 1 0 0 1 1 0 0 0 1 1 0 0 1 1 2 2 0 = 1 0 1 0 0 n t

Compressive Sensing 0. 1 0. 2 0 1 1 0 0 0 1 1 1 1 0 0 1 1 0 0 0 1 1 0 0 1 1 0. 1 13. 7 5. 8 0. 1 13. 9 0. 3 0. 7 0. 1 = 0. 2 6. 4 7. 3 1. 0 8. 2 0. 1 t 0. 1 0. 2 n

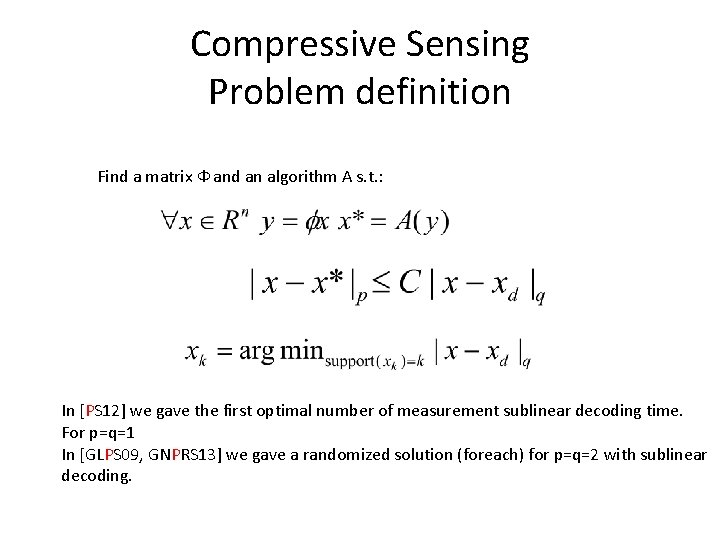
Compressive Sensing Problem definition Find a matrix Ф and an algorithm A s. t. : In [PS 12] we gave the first optimal number of measurement sublinear decoding time. For p=q=1 In [GLPS 09, GNPRS 13] we gave a randomized solution (foreach) for p=q=2 with sublinear decoding.

How Compressive Sensing help Massive Recommender Systems • Consider designing recommender system for web pages – Time a user examines a page is an implicit rating – Millions of users – Each user examines thousands of pages throughout the year – Hard to store and process the information

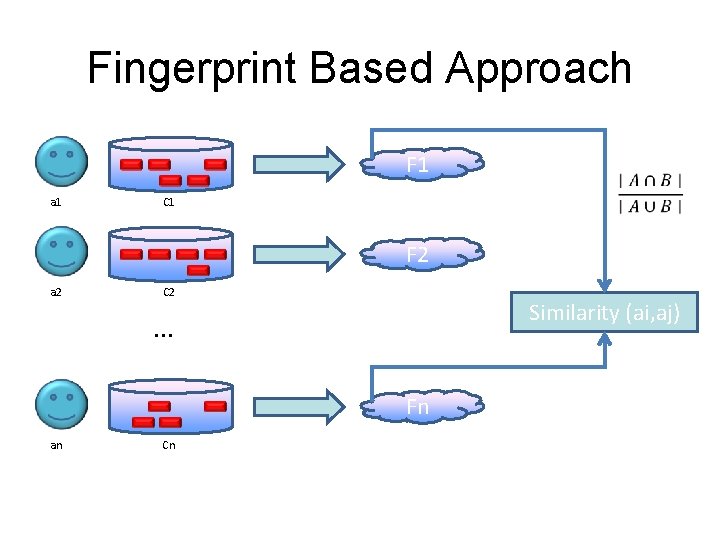
Fingerprint Based Approach F 1 a 1 C 1 F 2 a 2 C 2 Similarity (ai, aj) . . . Fn an Cn

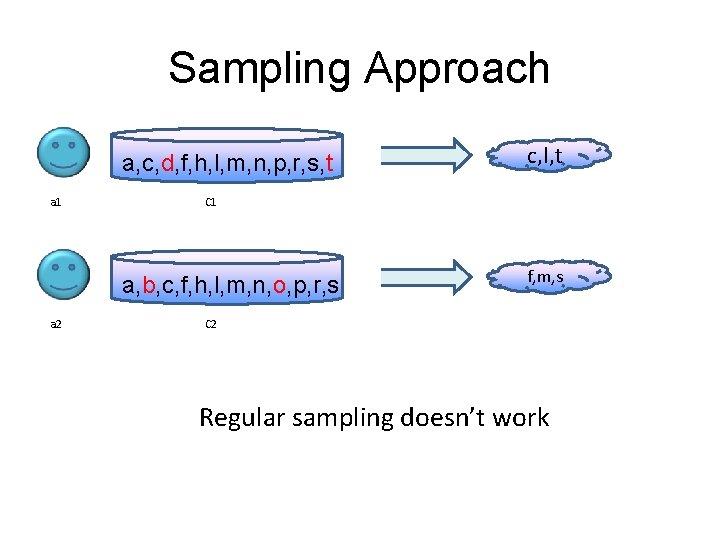
Sampling Approach a, c, d, f, h, l, m, n, p, r, s, t a 1 C 1 a, b, c, f, h, l, m, n, o, p, r, s a 2 c, l, t f, m, s C 2 Regular sampling doesn’t work

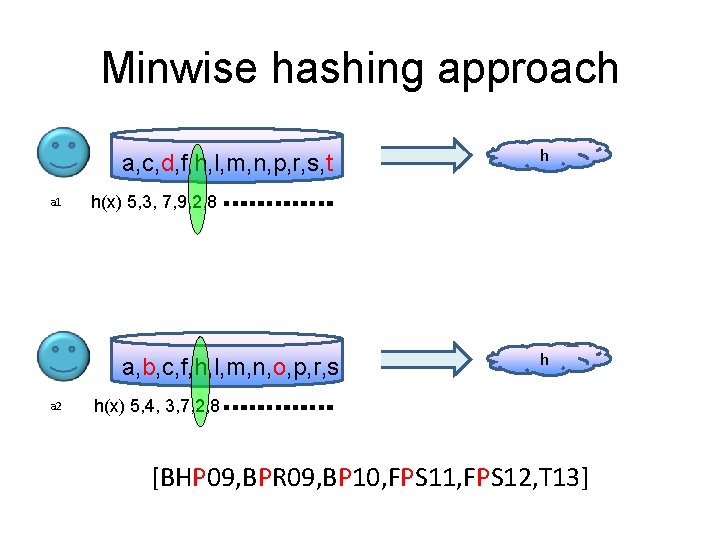
Minwise hashing approach a, c, d, f, h, l, m, n, p, r, s, t a 1 h(x) 5, 3, 7, 9, 2, 8 a, b, c, f, h, l, m, n, o, p, r, s a 2 h h h(x) 5, 4, 3, 7, 2, 8 [BHP 09, BPR 09, BP 10, FPS 11, FPS 12, T 13]

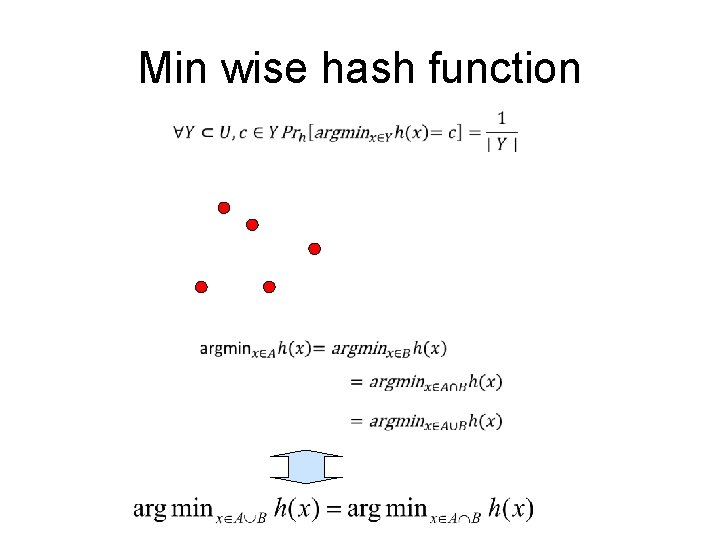
Min wise hash function

Min wise hash function

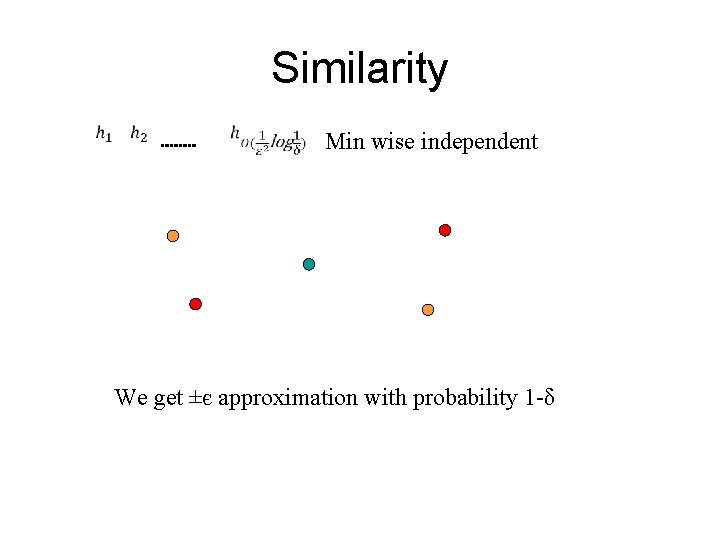
Similarity Min wise independent We get ±є approximation with probability 1 -δ
![Reducing sketching space BP 10 Instead of Additional pairwise independent hash It was discover Reducing sketching space [BP 10] Instead of Additional pairwise independent hash It was discover](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-38.jpg)
Reducing sketching space [BP 10] Instead of Additional pairwise independent hash It was discover independently by Ping Li and Christian Konig
![Reducing sketching space BP 10 Our algorithm estimates Reducing sketching space [BP 10] Our algorithm estimates](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-39.jpg)
Reducing sketching space [BP 10] Our algorithm estimates
![Reducing sketching space even farther BP 10 We usually interesting in the case that Reducing sketching space even farther [BP 10] We usually interesting in the case that](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-40.jpg)
Reducing sketching space even farther [BP 10] We usually interesting in the case that sets are very similar. Assume J>1 -t => p>1 -0. 5 t A B A-B 0 1 1 0 1 0 1 1 0 0 0 -1 0 0 0 CS 2 0 -2
![Reducing sketching space even farther BP 10 We usually interesting in the case that Reducing sketching space even farther [BP 10] We usually interesting in the case that](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-41.jpg)
Reducing sketching space even farther [BP 10] We usually interesting in the case that sets are very similar. Assume J>1 -t => p>1 -0. 5 t A B A xor B 0 1 1 0 1 0 1 1 0 0 0 CS 1 0 1 This give an improvement of
![Removing the min wise independent requirement BP 11 KNW 10 gave bits sketch Removing the min wise independent requirement [BP 11] • [KNW 10] gave bits sketch](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-42.jpg)
Removing the min wise independent requirement [BP 11] • [KNW 10] gave bits sketch for distinct count (F 0) • Their sketch is not linear – However given S(A) and S(B) one can calculate S(A+B) (that will give the size of the union)
![Removing the min wise independent requirement BP 11 Using F 2 instead of F Removing the min wise independent requirement [BP 11] Using F 2 instead of F](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-43.jpg)
Removing the min wise independent requirement [BP 11] Using F 2 instead of F 0 we managed to reduce the sketch size to Using more randomness we mange to remove factor

File sharing The naïve way

File sharing Torrent/Emule/Kazaa

File sharing Source: Clients: Coupon collector O(nlogn) In practice it could be 7 Gb instead 1 Gb

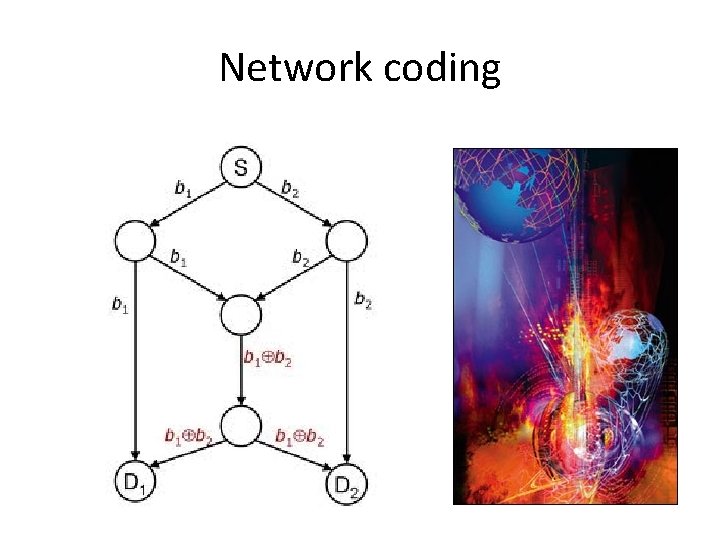
Network coding

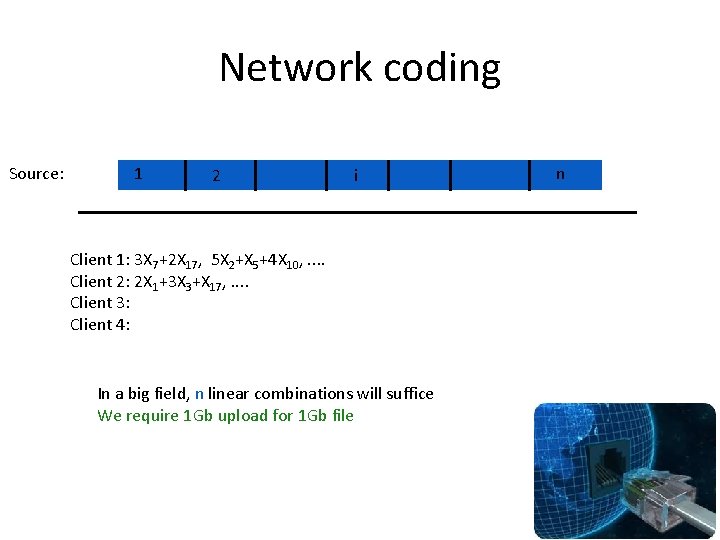
Network coding Source: 1 2 i Client 1: 3 X 7+2 X 17, 5 X 2+X 5+4 X 10, . . Client 2: 2 X 1+3 X 3+X 17, . . Client 3: Client 4: In a big field, n linear combinations will suffice We require 1 Gb upload for 1 Gb file n

Poison Torrent/Emule/Kaza

Signatures against poison 1 2 n i MD 5 Si . torrent file S 1 S 2. . . Sn We might receive poisoned packet But we won't forward it

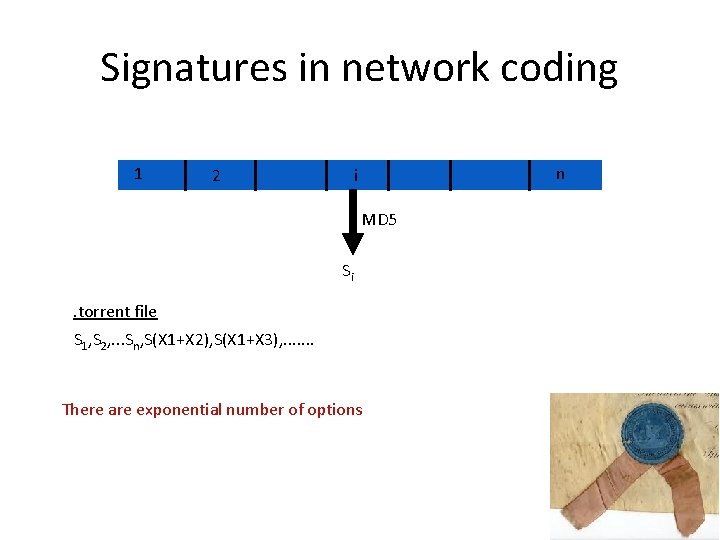
Signatures in network coding 1 2 n i MD 5 Si . torrent file S 1, S 2, . . . Sn, S(X 1+X 2), S(X 1+X 3), . . . . There are exponential number of options

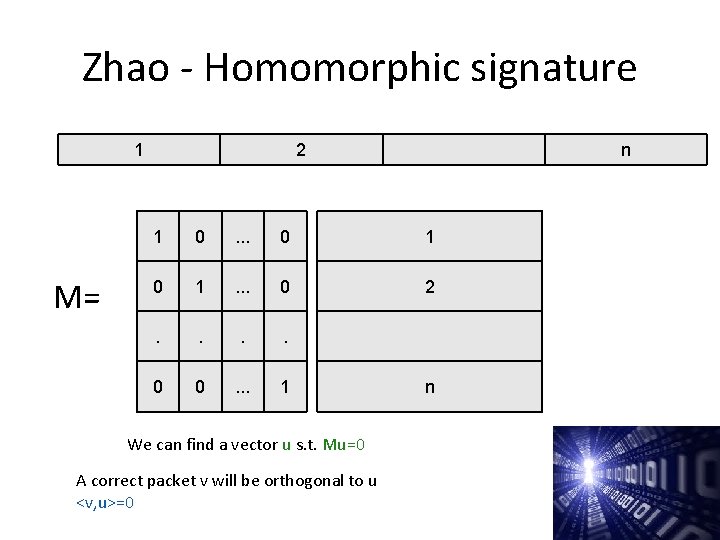
Zhao - Homomorphic signature 1 M= 2 n 1 0 . . . 0 1 . . . 0 2 . . 0 0 . . . 1 We can find a vector u s. t. Mu=0 A correct packet v will be orthogonal to u <v, u>=0 n

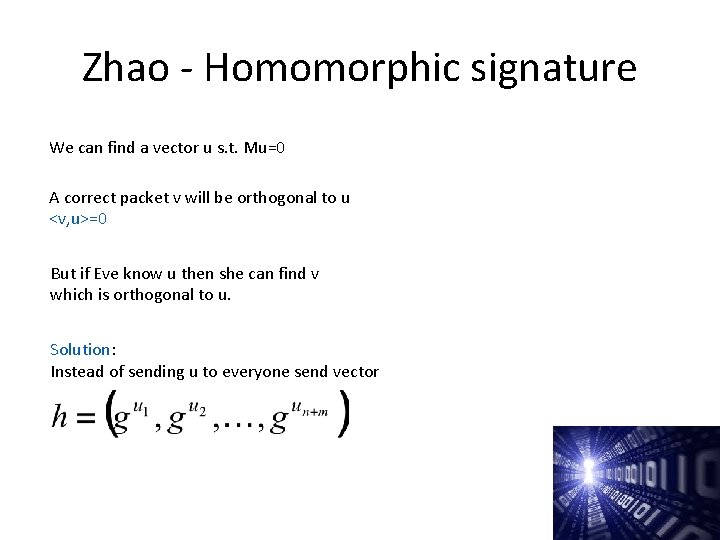
Zhao - Homomorphic signature We can find a vector u s. t. Mu=0 A correct packet v will be orthogonal to u <v, u>=0 But if Eve know u then she can find v which is orthogonal to u. Solution: Instead of sending u to everyone send vector

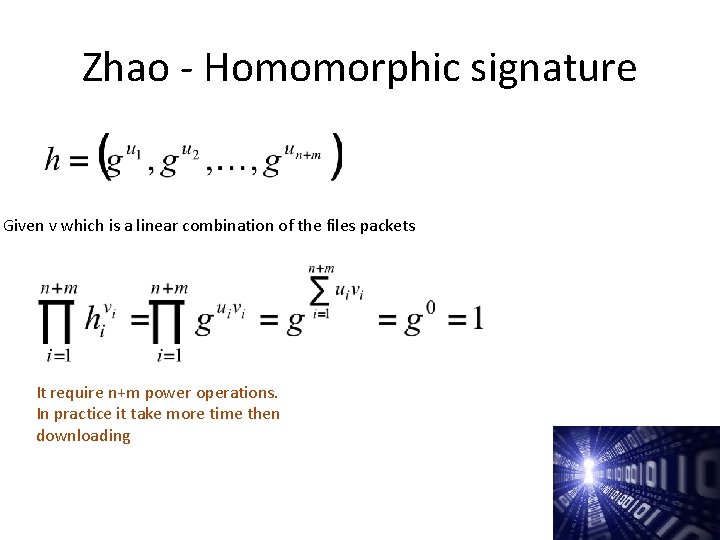
Zhao - Homomorphic signature Given v which is a linear combination of the files packets It require n+m power operations. In practice it take more time then downloading
![Selective verification PW 12 Packeti Si If we have both signatures we can choose Selective verification [PW 12] Packeti S'i If we have both signatures we can choose](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-55.jpg)
Selective verification [PW 12] Packeti S'i If we have both signatures we can choose randomly which to check S''i

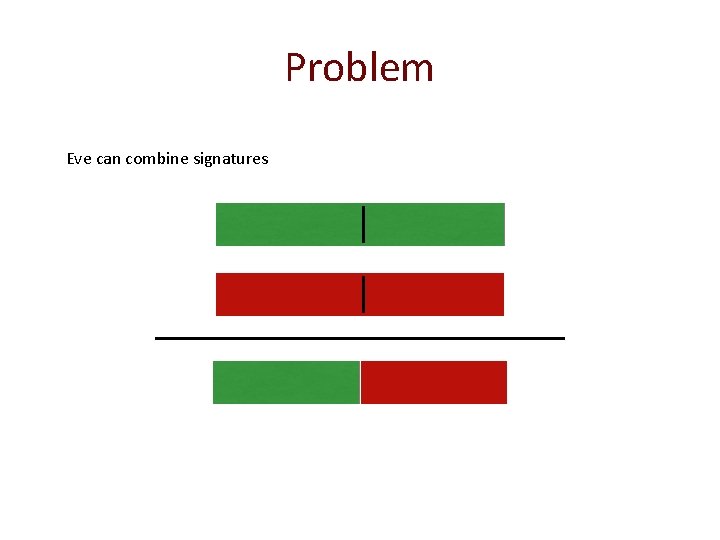
Problem Eve can combine signatures

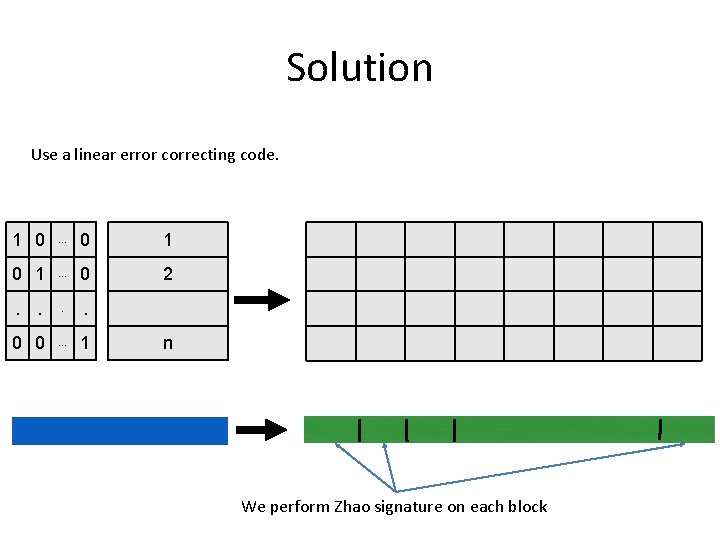
Solution Use a linear error correcting code. 1 0 . . . 0 1 . . . 0 2 . . 0 0 . . . 1 n We perform Zhao signature on each block

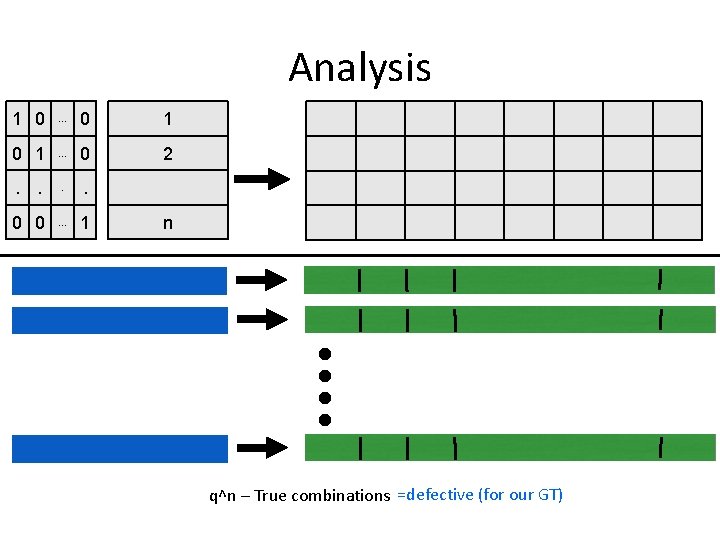
Analysis 1 0 . . . 0 1 . . . 0 2 . . 0 0 . . . 1 n q^n – True combinations =defective (for our GT)
![Analysis 1 nm 2 dn Prone block pass the testqnqdnqd1n Prr2 out of r Analysis 1 n+m 2 dn Pr[one block pass the test]<qn/qdn=q-(d-1)n Pr[r/2 out of r](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-59.jpg)
Analysis 1 n+m 2 dn Pr[one block pass the test]<qn/qdn=q-(d-1)n Pr[r/2 out of r pass the test]< 2 rq-(d-1)r/2 r
![Analysis 1 nm 2 dn Prone block pass the testqnqdnqd1n Prr2 out of r Analysis 1 n+m 2 dn Pr[one block pass the test]<qn/qdn=q-(d-1)n Pr[r/2 out of r](https://slidetodoc.com/presentation_image_h2/fc8824b6748dbd859ea9eff528848cfe/image-60.jpg)
Analysis 1 n+m 2 dn Pr[one block pass the test]<qn/qdn=q-(d-1)n Pr[r/2 out of r pass the test]< 2 rq-(d-1)r/2 Using union bound: the probability that a bad packet exist is bounded by q(n+m)+r/log q-(d-1)nr In practice we improve Zhao signature by a factor of 60. r

Conclusion • Group testing/Compressive sensing is very effective tool. • We improved both construction and achieved sublinear decoding time. • Surprising important applications.