Group Sharing Wordle www wordle net What is
















- Slides: 40

Group Sharing

Wordle www. wordle. net

What is antibiotic resistance? • Antibiotic resistance often confused with virulence • Virulence refers to factors that enable a bacterium to attach to host cells, invade tissue, avoid the immune system, form biofilms and establish an infection • Antibiotic resistance refers to the ability of a bacterium to grow in the presence of antibiotics

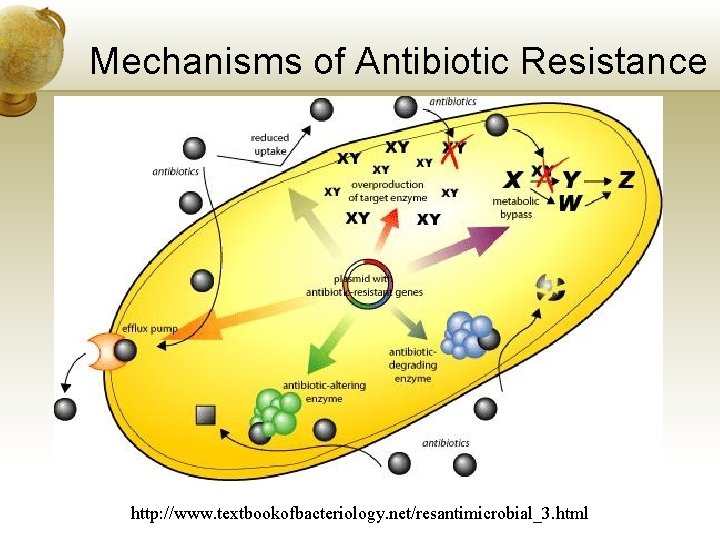
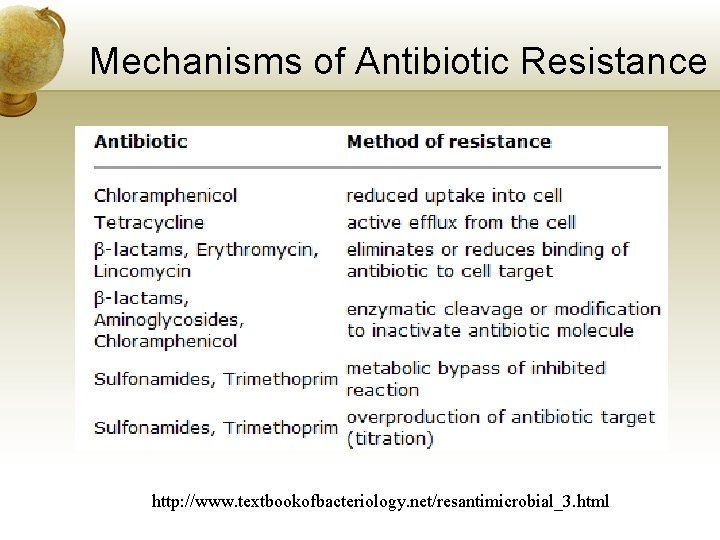
Mechanisms of Antibiotic Resistance http: //www. textbookofbacteriology. net/resantimicrobial_3. html

Mechanisms of Antibiotic Resistance http: //www. textbookofbacteriology. net/resantimicrobial_3. html

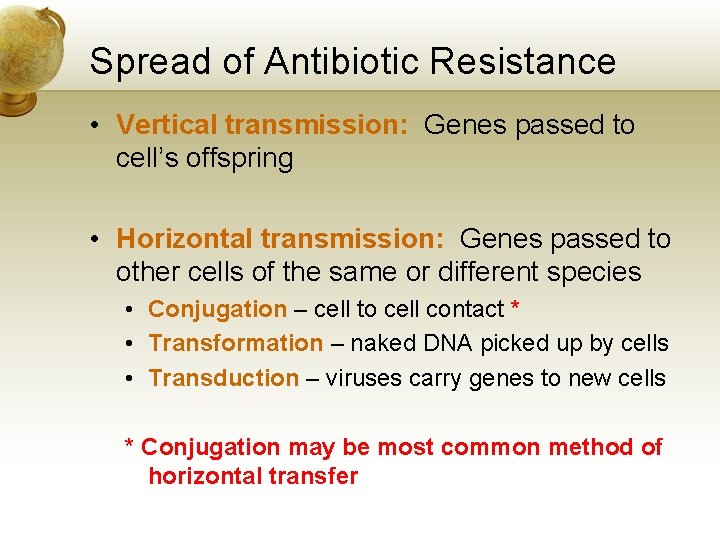
Spread of Antibiotic Resistance • Vertical transmission: Genes passed to cell’s offspring • Horizontal transmission: Genes passed to other cells of the same or different species • Conjugation – cell to cell contact * • Transformation – naked DNA picked up by cells • Transduction – viruses carry genes to new cells * Conjugation may be most common method of horizontal transfer

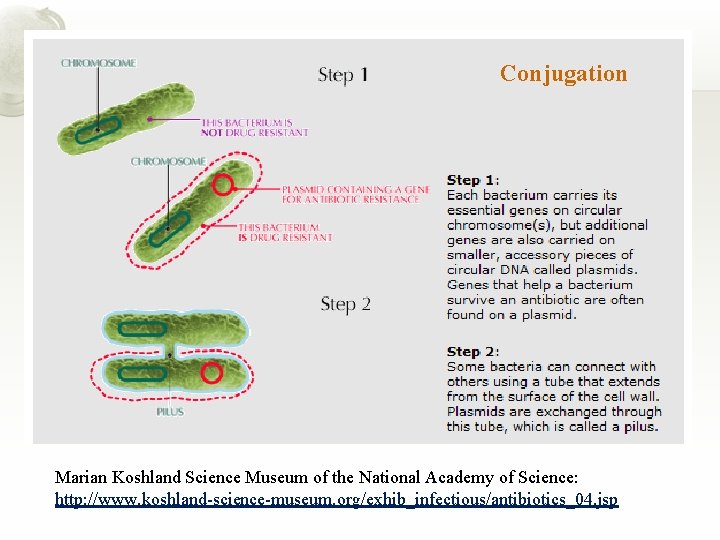
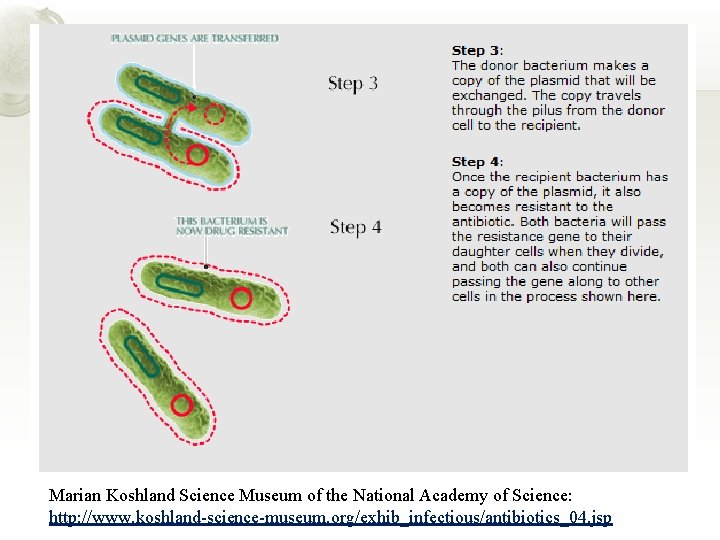
Conjugation Marian Koshland Science Museum of the National Academy of Science: http: //www. koshland-science-museum. org/exhib_infectious/antibiotics_04. jsp

Marian Koshland Science Museum of the National Academy of Science: http: //www. koshland-science-museum. org/exhib_infectious/antibiotics_04. jsp

What is the origin of resistance genes? • In nature antibiotics function at low concentrations in metabolic and regulatory pathways • Antibiotic resistance genes coevolved with these “antibiotics” • In nature resistance genes may function in detoxification of metabolic intermediates; inhibition of virulence factors; regulation of signal trafficking • In clinical settings, the high concentrations of antibiotics apply selective pressure and the most important function of the genes is for resistance to the deadly effects of the antibiotic

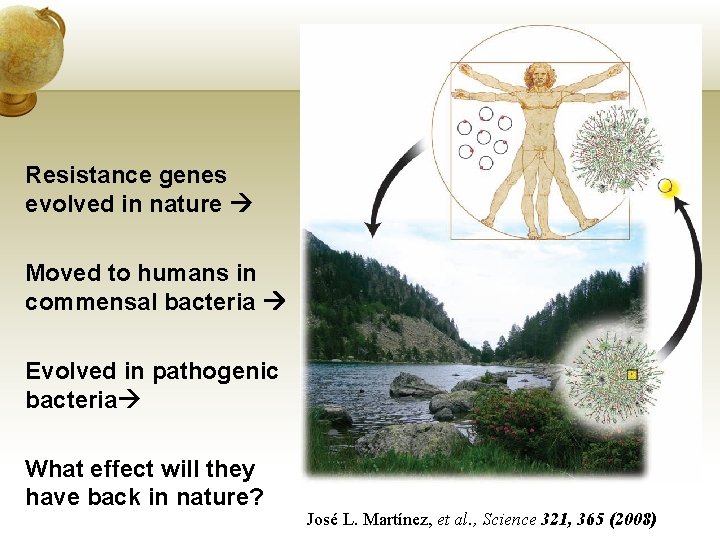
Resistance genes evolved in nature Moved to humans in commensal bacteria Evolved in pathogenic bacteria What effect will they have back in nature? José L. Martínez, et al. , Science 321, 365 (2008)

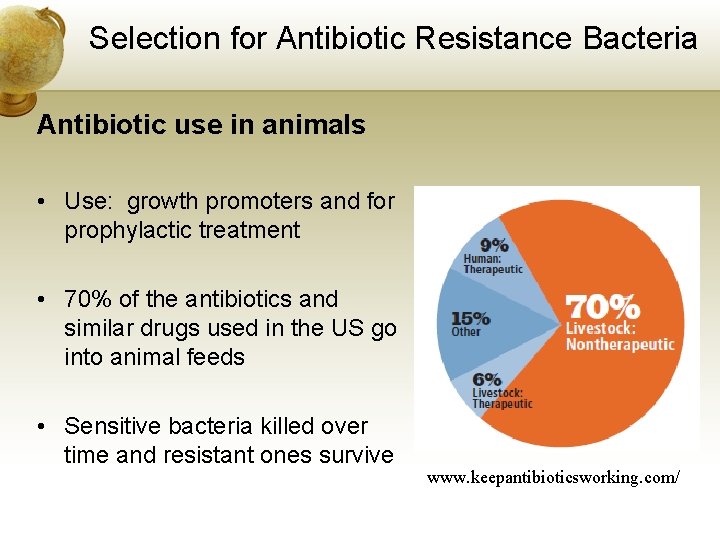
Selection for Antibiotic Resistance Bacteria Antibiotic use in animals • Use: growth promoters and for prophylactic treatment • 70% of the antibiotics and similar drugs used in the US go into animal feeds • Sensitive bacteria killed over time and resistant ones survive www. keepantibioticsworking. com/

Selection for Antibiotic Resistance Bacteria Antibiotic use in humans - concerns • Prescribing bacterial antibiotics for viruses • Prescribing the wrong drug or not checking susceptibility of organism • Not completing the whole course of drugs • Biofilms at infection site protecting bacteria

Over the Counter Antibiotics • Self-prescribing and taking meds for too few days • Antibiotics available in many countries • 6/6/2010

http: //antibiotics. withoutaprescription. net/

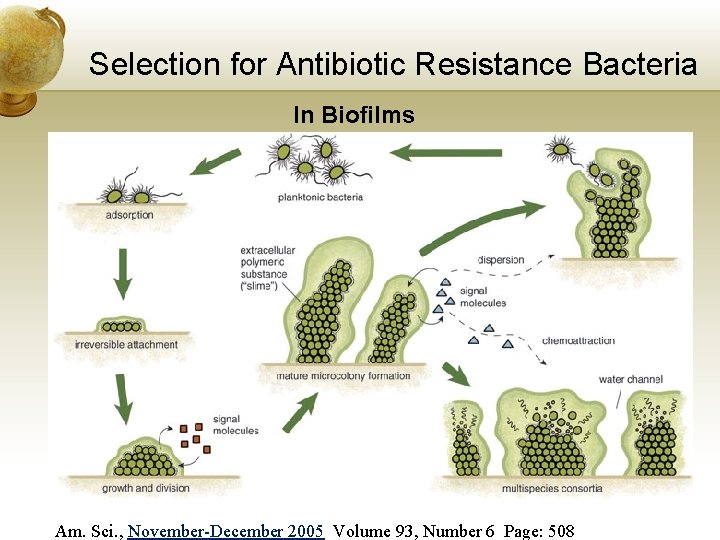
Selection for Antibiotic Resistance Bacteria In Biofilms Am. Sci. , November-December 2005 Volume 93, Number 6 Page: 508

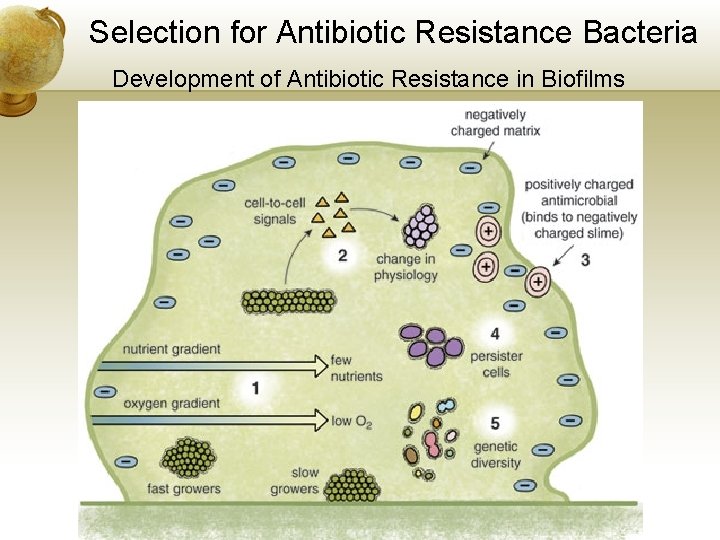
Selection for Antibiotic Resistance Bacteria Development of Antibiotic Resistance in Biofilms

Biofilms…. . are hiding in plain sight. • Human sites: middle ear, teeth, intestines, infected lungs • Conjugation and transformation occur in biofilms spread of resistance genes

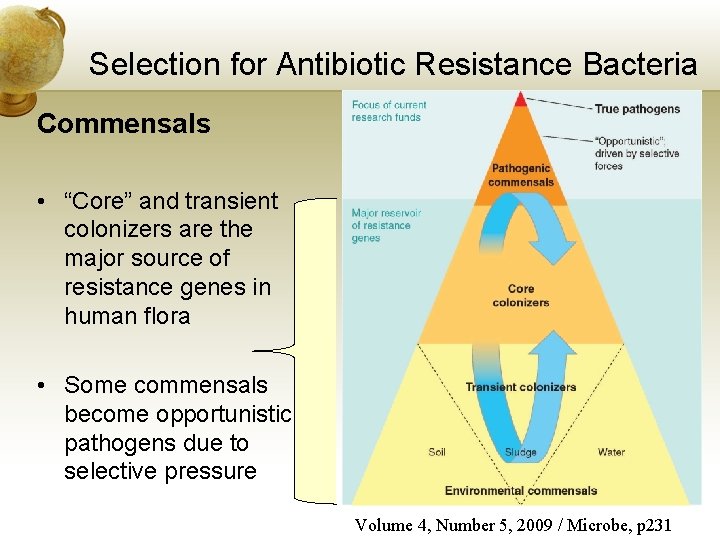
Selection for Antibiotic Resistance Bacteria Commensals • “Core” and transient colonizers are the major source of resistance genes in human flora • Some commensals become opportunistic pathogens due to selective pressure Volume 4, Number 5, 2009 / Microbe, p 231

Selection for Antibiotic Resistance Bacteria Commensals • Alliance for the Prudent Use of Antibiotics has started a project of evaluating and tracking antibiotic resistance genes in nature since data is scattered and incomplete • Project called ROAR – Reservoirs of Antibiotic Resistance Network • • Collect & analyze genetic data (using literature) Modeling Encouraging funding and research for this work ROAR

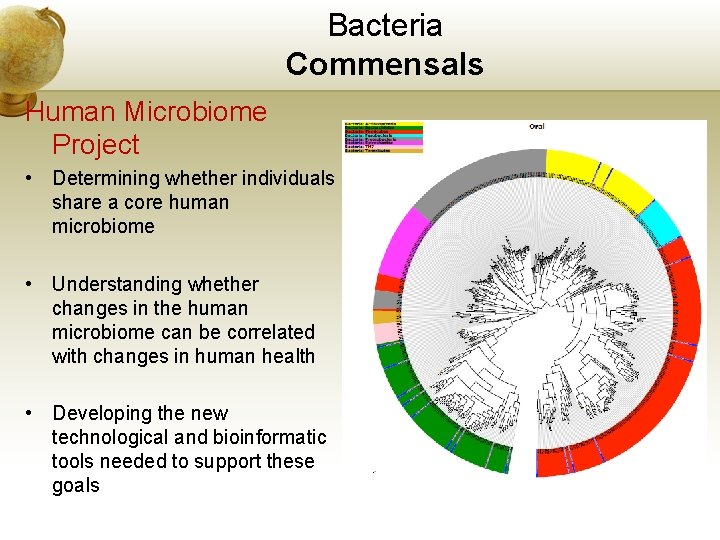
Bacteria Commensals Human Microbiome Project • Determining whether individuals share a core human microbiome • Understanding whether changes in the human microbiome can be correlated with changes in human health • Developing the new technological and bioinformatic tools needed to support these goals

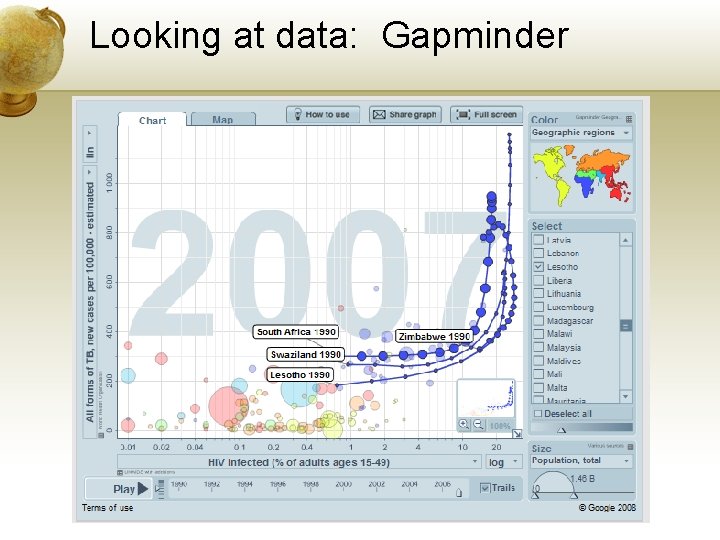
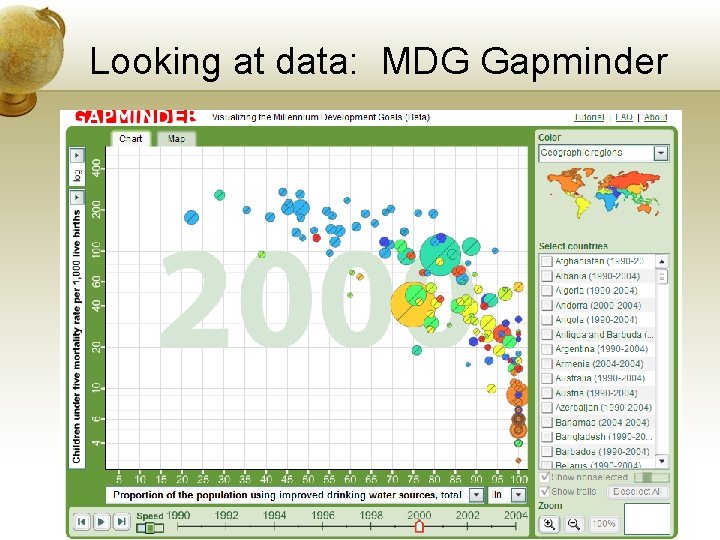
Looking at data: Gapminder

Looking at data: Worldmapper • www. worldmapper. org • Can download Excel file with data and notes • Can compare with a territory or pop. map

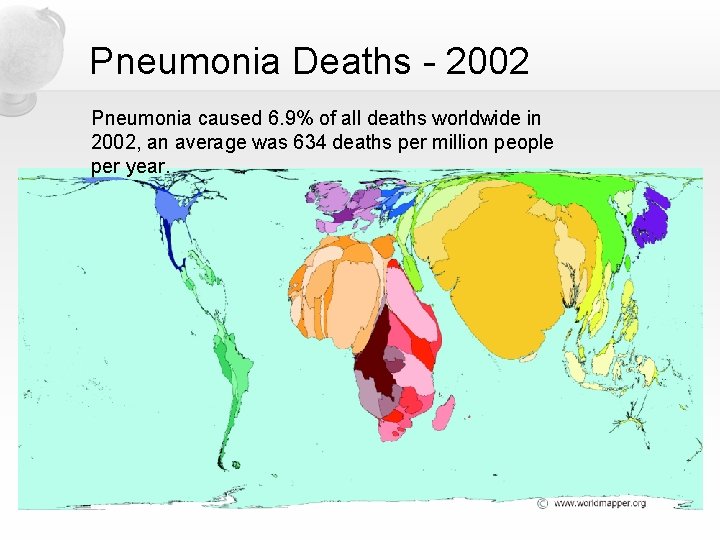
Pneumonia Deaths - 2002 Pneumonia caused 6. 9% of all deaths worldwide in 2002, an average was 634 deaths per million people per year.

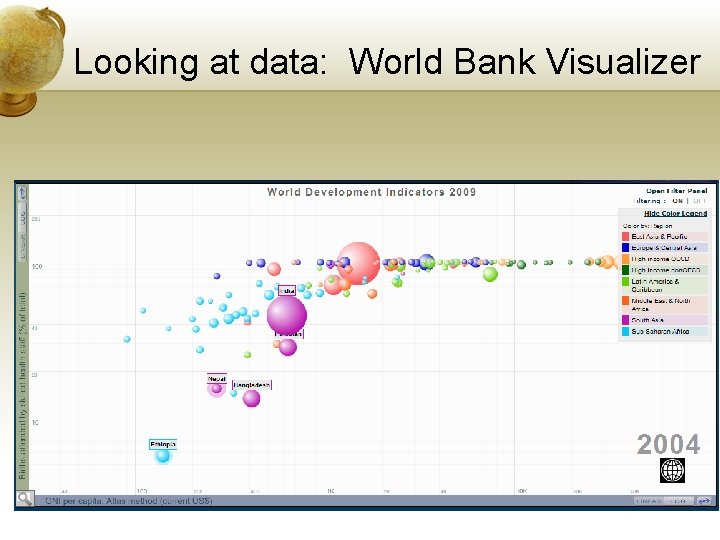
Looking at data: World Bank Visualizer

Looking at data: MDG Gapminder

Sources of data These sites provide data sets- • • • World Bank Gapminder data Worldmapper data WHO UN divisions

Collecting and Analyzing Data • ESAC – European Surveillance of Antimicrobial Consumption - database • EARSS – European Antimicrobial Resistance Surveillance System - database • Example: Examine the relationship between antibiotic consumption and resistance • % Streptococcus pneumoniae that are antibiotic resistant • % Staphylococcus aureus that are MRSA (methicillin resistant)

Streptococcus pneumoniae • Part of normal flora of upper respiratory system • Causes pneumonia, ear infections, sinusitis • Leading cause of invasive bacterial disease in children and the elderly

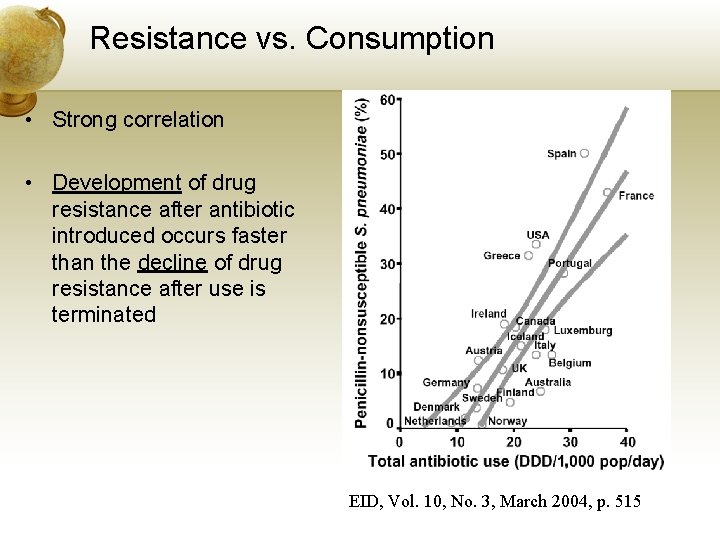
Resistance vs. Consumption • Strong correlation • Development of drug resistance after antibiotic introduced occurs faster than the decline of drug resistance after use is terminated EID, Vol. 10, No. 3, March 2004, p. 515

Working with data • Google Docs Motion Gadget • Create data in Google Docs spreadsheet • Insert Motion Gadget • Choose the range for data • http: //spreadsheets. google. com/ccc? key=0 As. QAokb_R_yd. HFMc. VFuenp. Va. UNQSXJGLWZFVUh. KVmc&hl=en • Data set prepared in Google Docs: • Antibiotic consumption • % Staphylococcus aureus that are MRSA • % Streptococcus pneumoniae that are resistant to penicillin

Things to consider • What kind of questions can you ask? • What happens with missing data? • Does seeing at a correlation demonstrate causality?

THE END




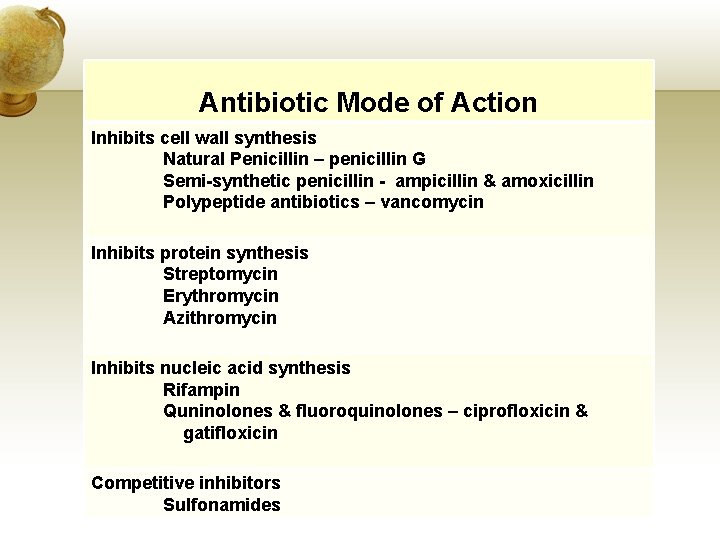
Antibiotic Mode of Action Inhibits cell wall synthesis Natural Penicillin – penicillin G Semi-synthetic penicillin - ampicillin & amoxicillin Polypeptide antibiotics – vancomycin Inhibits protein synthesis Streptomycin Erythromycin Azithromycin Inhibits nucleic acid synthesis Rifampin Quninolones & fluoroquinolones – ciprofloxicin & gatifloxicin Competitive inhibitors Sulfonamides


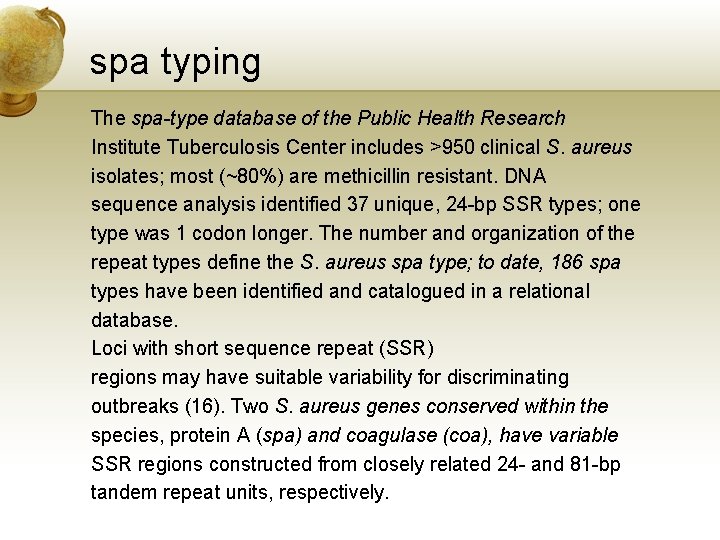
spa typing The spa-type database of the Public Health Research Institute Tuberculosis Center includes >950 clinical S. aureus isolates; most (~80%) are methicillin resistant. DNA sequence analysis identified 37 unique, 24 -bp SSR types; one type was 1 codon longer. The number and organization of the repeat types define the S. aureus spa type; to date, 186 spa types have been identified and catalogued in a relational database. Loci with short sequence repeat (SSR) regions may have suitable variability for discriminating outbreaks (16). Two S. aureus genes conserved within the species, protein A (spa) and coagulase (coa), have variable SSR regions constructed from closely related 24 - and 81 -bp tandem repeat units, respectively.

• We now know more about the mechanism by which bacteria develop antibiotic resistance. Strains of methicillin-resistant S. aureus have developed through acquisition of mobile genetic elements which carry the mec. A gene, which encodes a protein (penicillin-binding protein 2 a) that penicillin does not bind or attach to. The effectiveness of penicillin antibiotics relies on the ability to bind to the surface of the bacteria. The mec. A gene is carried on a mobile genetic element called a cassette (SCCmec cassette). A genetic cassette may carry a number of genetic elements, much like a compact disc holds more than one song – in the case of MRSA, the cassette carries the mec. A gene, and depending on the strain of S. aureus may also carry other resistance factors that provide protection against other antibiotics, as well as a variety of virulence factors.

• Currently, there are 4 recognized clones or types of SCCmec - I through IV, with SCCmec IV being the most common. The structure of the mec. A genes are similar, but the gene structure is significantly different than the hospital-acquired strains. Because of the differences in mec. A gene structure and cassette components, hospital-acquired strains tend to be resistant to multiple antibiotics, and are associated more often with surgical wound infections, urinary tract infections, bloodstream infections, and pneumonia. Community-acquired strains do not share the same antibiotic resistance patterns as the strains circulating in the hospital, and tend to cause infections associated with methicillin susceptible strains of S. aureus in the community – skin infections (pimples, boils, impetigo, infected cuts, etc. ), but can also cause more serious infections like pneumonia. • Read more at Suite 101: Drug Resistant Bacteria: Relationship Between Community and Hospital Acquired Infections http: //microbiology. suite 101. com/article. cfm/mrsa__camrsa_an d_hamrsa#ixzz 0 qb. YDM 6 r. U
 Http://wordle
Http://wordle Wordle.net
Wordle.net Wordle.net
Wordle.net Wordle net
Wordle net Literacy wordle
Literacy wordle Http://www.wordle.net/
Http://www.wordle.net/ Parle wordle
Parle wordle Http://www.wordle.net/create
Http://www.wordle.net/create Www.wordle.net
Www.wordle.net Achmed lach net
Achmed lach net Ado.net vb.net
Ado.net vb.net J.b. boda net worth
J.b. boda net worth Group net for plan members
Group net for plan members Wordle solver
Wordle solver Quark physics wordle
Quark physics wordle Wordle poem
Wordle poem Wordle mandarin
Wordle mandarin Wordle en ligne
Wordle en ligne Wordle 2/1
Wordle 2/1 Wordle starters
Wordle starters Wordle orge
Wordle orge Wordle 205x
Wordle 205x Wordle
Wordle Wordle
Wordle Ekg lead wall
Ekg lead wall Customer service wordle
Customer service wordle Cristiano ronaldo heart disease
Cristiano ronaldo heart disease Wordle of rhe day
Wordle of rhe day Ordsky program
Ordsky program Lyrics in my heart there rings a melody
Lyrics in my heart there rings a melody Wordle 204 3/6
Wordle 204 3/6 Wordle 209
Wordle 209 Wordle #9
Wordle #9 Teamwork wordle
Teamwork wordle Social psychology chapter 13
Social psychology chapter 13 Y = a(b)^x
Y = a(b)^x Anova within group and between group
Anova within group and between group Primary group
Primary group Jrcptb
Jrcptb Thermal stability of group 1 nitrates
Thermal stability of group 1 nitrates Amino group and carboxyl group
Amino group and carboxyl group