Great Questions and Participation in the Last Two






- Slides: 49

Great Questions and Participation in the Last Two Classes!!!! We gave each of you maximum points! Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects

Homework and Presentation Deadlines • Please do not delay handing in the homeworks • Please do not wait until the last minute to finalize your presentation before meeting with us… 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 2

“Natively unfolded proteins” and what the biophysical methods can report on them Judith Klein-Seetharaman Co-Course Director jks 33@pitt. edu Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects

Lecture Overview • Natively unfolded proteins • Brief circular dichroism tutorial • Example CD: Is the transducer natively unfolded? • Brief intro to methods for “seeing” molecules • Example SANS: Is the transducer natively unfolded? • Theories on solvent effects • Alternative methods for “seeing” molecules: AFM, Cryo. EM outlook 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 4

Lecture Overview • Natively unfolded proteins • Brief circular dichroism tutorial • Example CD: Is the transducer natively unfolded? • Brief intro to methods for “seeing” molecules • Example SANS: Is the transducer natively unfolded? • Theories on solvent effects • Alternative method for “seeing” molecules: AFM 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 5

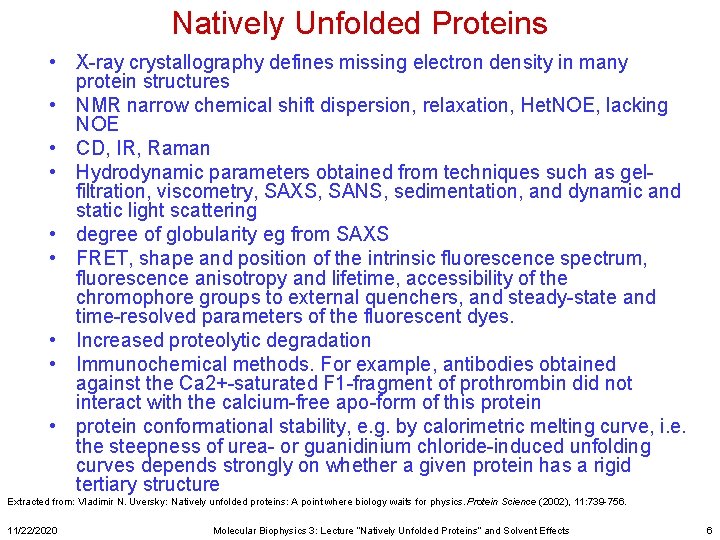
Natively Unfolded Proteins • X-ray crystallography defines missing electron density in many protein structures • NMR narrow chemical shift dispersion, relaxation, Het. NOE, lacking NOE • CD, IR, Raman • Hydrodynamic parameters obtained from techniques such as gelfiltration, viscometry, SAXS, SANS, sedimentation, and dynamic and static light scattering • degree of globularity eg from SAXS • FRET, shape and position of the intrinsic fluorescence spectrum, fluorescence anisotropy and lifetime, accessibility of the chromophore groups to external quenchers, and steady-state and time-resolved parameters of the fluorescent dyes. • Increased proteolytic degradation • Immunochemical methods. For example, antibodies obtained against the Ca 2+-saturated F 1 -fragment of prothrombin did not interact with the calcium-free apo-form of this protein • protein conformational stability, e. g. by calorimetric melting curve, i. e. the steepness of urea- or guanidinium chloride-induced unfolding curves depends strongly on whether a given protein has a rigid tertiary structure Extracted from: Vladimir N. Uversky: Natively unfolded proteins: A point where biology waits for physics. Protein Science (2002), 11: 739 -756. 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 6

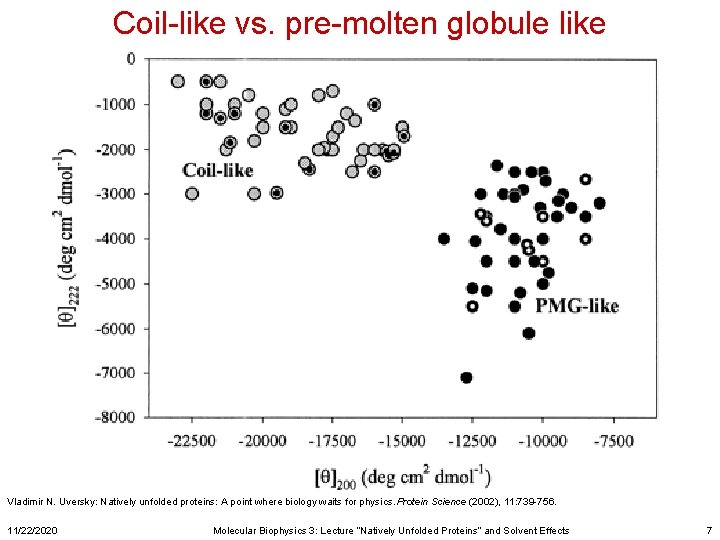
Coil-like vs. pre-molten globule like Vladimir N. Uversky: Natively unfolded proteins: A point where biology waits for physics. Protein Science (2002), 11: 739 -756. 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 7

Prediction of Disorder • PONDR – Neural network from sequence features • SVM • others 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 8

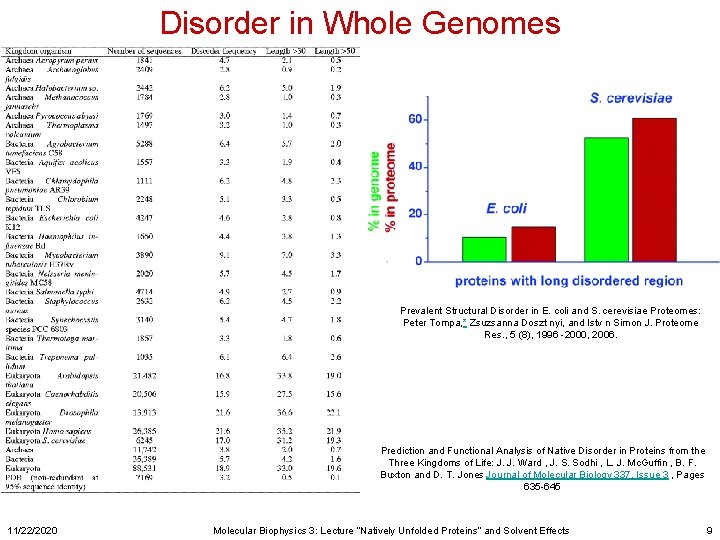
Disorder in Whole Genomes Prevalent Structural Disorder in E. coli and S. cerevisiae Proteomes: Peter Tompa, * Zsuzsanna Doszt nyi, and Istv n Simon J. Proteome Res. , 5 (8), 1996 -2000, 2006. Prediction and Functional Analysis of Native Disorder in Proteins from the Three Kingdoms of Life: J. J. Ward , J. S. Sodhi , L. J. Mc. Guffin , B. F. Buxton and D. T. Jones Journal of Molecular Biology 337, Issue 3 , Pages 635 -645 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 9

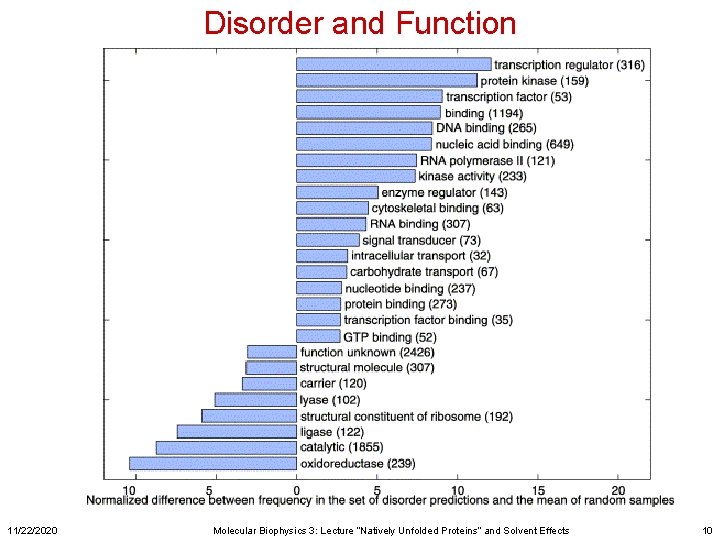
Disorder and Function 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 10

Lecture Overview • Natively unfolded proteins • Brief circular dichroism tutorial • Example CD: Is the transducer natively unfolded? • Brief intro to methods for “seeing” molecules • Example SANS: Is the transducer natively unfolded? • Theories on solvent effects • Alternative method for “seeing” molecules: AFM 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 11

Circular Dichroism Tutorial • • • Remind you of what CD is What data do you get typically? How do you analyze it? Limitations Applications to study of dynamics and biomolecular interactions • Outline of the homework 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 12

Objectives of this Tutorial • • • Remind you of what CD is What data do you get typically? How do you analyze it? Limitations Applications to study of dynamics and biomolecular interactions • Outline of the homework 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 13

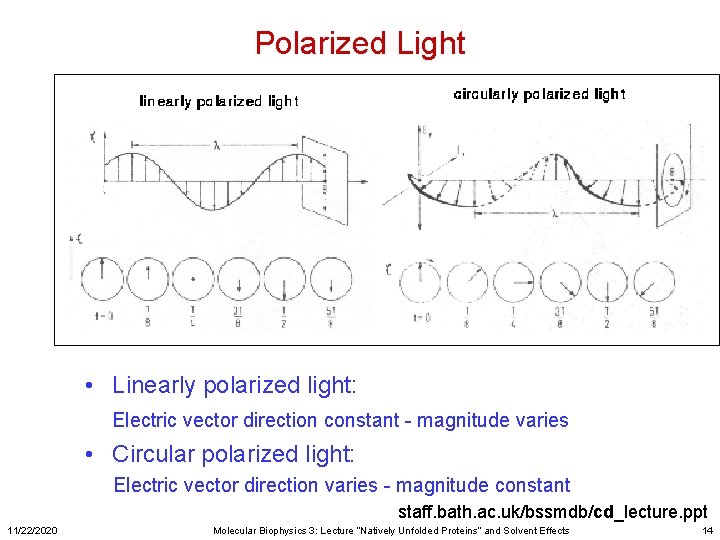
Polarized Light Crystals dark Crystals light • Linearly polarized light: Electric vector direction constant - magnitude varies • Circular polarized light: Electric vector direction varies - magnitude constant staff. bath. ac. uk/bssmdb/cd_lecture. ppt 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 14

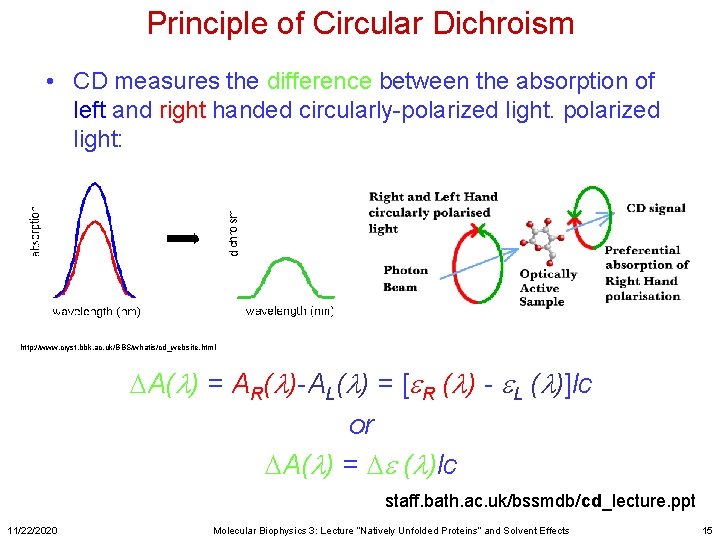
Principle of Circular Dichroism • CD measures the difference between the absorption of left and right handed circularly-polarized light: http: //www. cryst. bbk. ac. uk/BBS/whatis/cd_website. html DA(l) = AR(l)-AL(l) = [e. R (l) - e. L (l)]lc or DA(l) = De (l)lc staff. bath. ac. uk/bssmdb/cd_lecture. ppt 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 15

Objectives of this Tutorial • • • Remind you of what CD is What data do you get typically? How do you analyze it? Limitations Applications to study of dynamics and biomolecular interactions • Outline of the homework 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 16

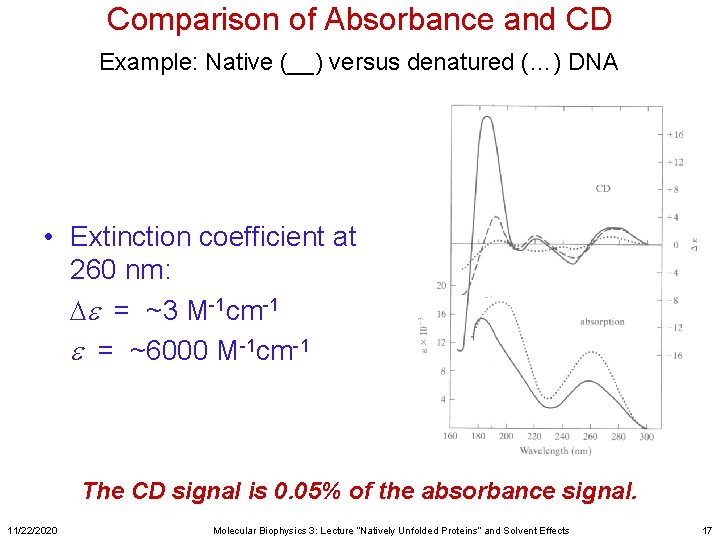
Comparison of Absorbance and CD Example: Native (__) versus denatured (…) DNA • Extinction coefficient at 260 nm: De = ~3 M-1 cm-1 e = ~6000 M-1 cm-1 The CD signal is 0. 05% of the absorbance signal. 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 17

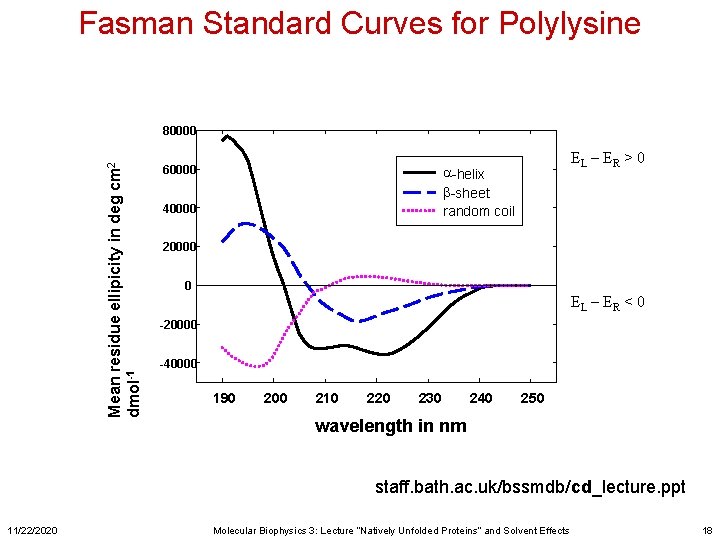
Fasman Standard Curves for Polylysine Mean residue ellipicity in deg cm 2 dmol-1 80000 60000 EL – ER > 0 a-helix b-sheet random coil 40000 20000 0 EL – ER < 0 -20000 -40000 190 200 210 220 230 240 250 wavelength in nm staff. bath. ac. uk/bssmdb/cd_lecture. ppt 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 18

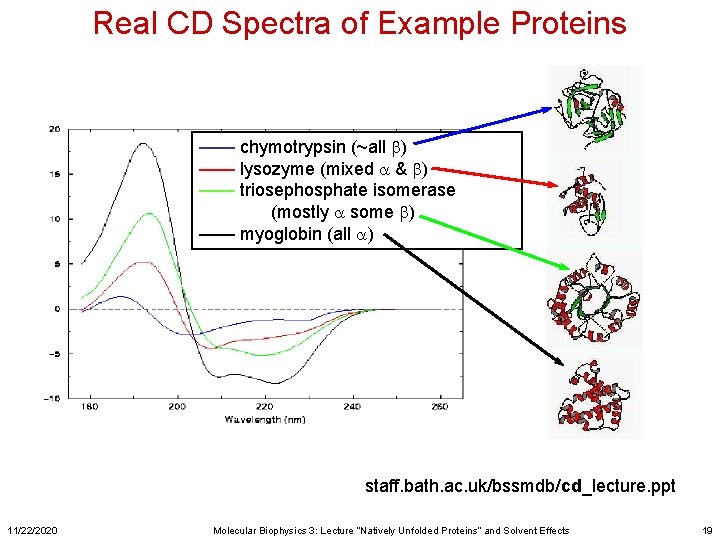
Real CD Spectra of Example Proteins —— chymotrypsin (~all b) —— lysozyme (mixed a & b) —— triosephosphate isomerase (mostly a some b) —— myoglobin (all a) staff. bath. ac. uk/bssmdb/cd_lecture. ppt 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 19

Objectives of this Tutorial • • • Remind you of what CD is What data do you get typically? How do you analyze it? Limitations Applications to study of dynamics and biomolecular interactions • Outline of the homework 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 20

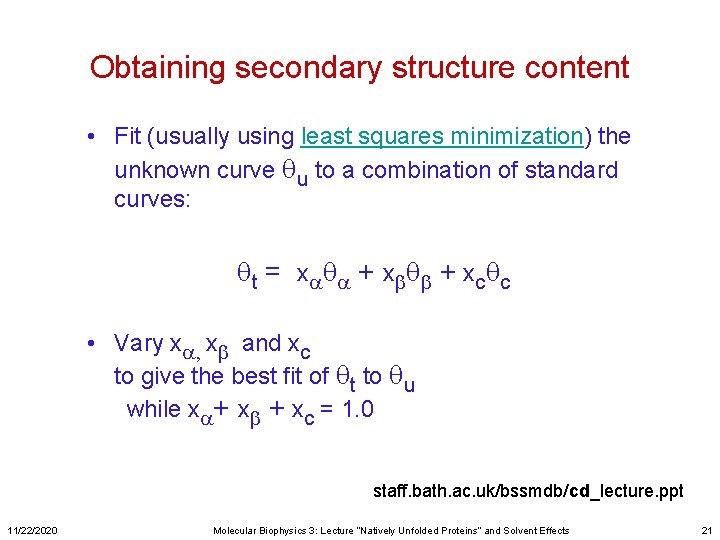
Obtaining secondary structure content • Fit (usually using least squares minimization) the unknown curve qu to a combination of standard curves: qt = xaqa + xbqb + xcqc • Vary xa, xb and xc to give the best fit of qt to qu while xa+ xb + xc = 1. 0 staff. bath. ac. uk/bssmdb/cd_lecture. ppt 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 21

Available methods • Check program descriptions on package websites: 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 22

CDPro Webinterface If you want to run the program within the webbrowser click Read. Me 11/22/2020 If you want to download the program to a PC, click “CDPro. zip” Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 23

Objectives of this Tutorial • • • Remind you of what CD is What data do you get typically? How do you analyze it? Limitations Applications to study of dynamics and biomolecular interactions • Outline of the homework 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 24

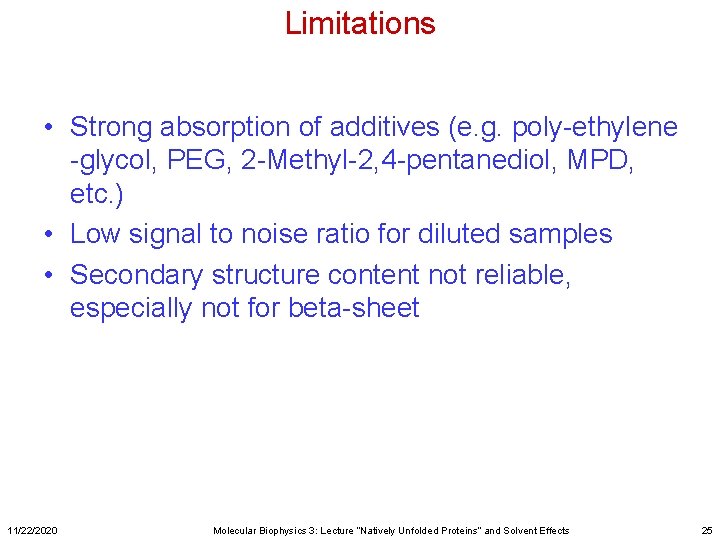
Limitations • Strong absorption of additives (e. g. poly-ethylene -glycol, PEG, 2 -Methyl-2, 4 -pentanediol, MPD, etc. ) • Low signal to noise ratio for diluted samples • Secondary structure content not reliable, especially not for beta-sheet 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 25

Objectives of this Tutorial • • • Remind you of what CD is What data do you get typically? How do you analyze it? Limitations Applications to study of dynamics and biomolecular interactions • Outline of the homework 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 26

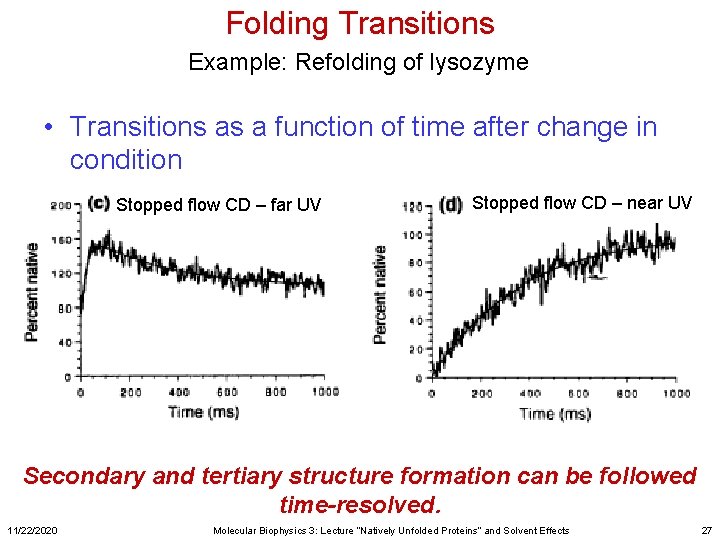
Folding Transitions Example: Refolding of lysozyme • Transitions as a function of time after change in condition Stopped flow CD – far UV Stopped flow CD – near UV Secondary and tertiary structure formation can be followed time-resolved. 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 27

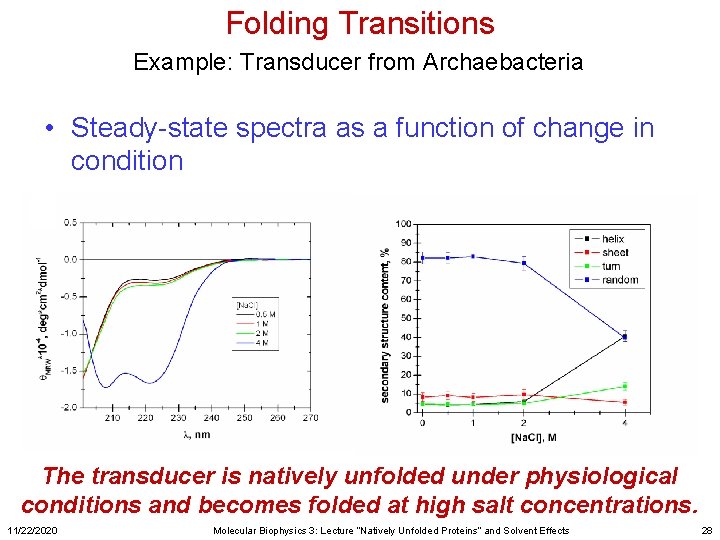
Folding Transitions Example: Transducer from Archaebacteria • Steady-state spectra as a function of change in condition The transducer is natively unfolded under physiological conditions and becomes folded at high salt concentrations. 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 28

Links Online and downloadable analysis tools: • Dichroweb www. cryst. bbk. ac. uk/cdweb/html/ • CDPro analysis package http: //lamar. colostate. edu/~sreeram/CDPro/ Tutorials: • Lecture similar to this one staff. bath. ac. uk/bssmdb/cd_lecture. ppt • Animation of polarized light http: //www. enzim. hu/~szia/cddemo/edemo 0. htm 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 29

Objectives of this Tutorial • • • Remind you of what CD is What data do you get typically? How do you analyze it? Limitations Applications to study of dynamics and biomolecular interactions • Outline of the homework 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 30

Homework: CD Analysis Step 1 Use the CDPro package to analyze primary CD data of protein X. http: //lamar. colostate. edu/~sreeram/CDPro/main. html Original data: Convert the two raw data files into files that are readable for the CDpro program by using CRDATA. exe Column 1: wavelength (should start from longer wavelength, e. g. 200 nm to 100 nm) Column 2: mean residue ellipticity (not molar ellipticity, teta) Separated by tab 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 31

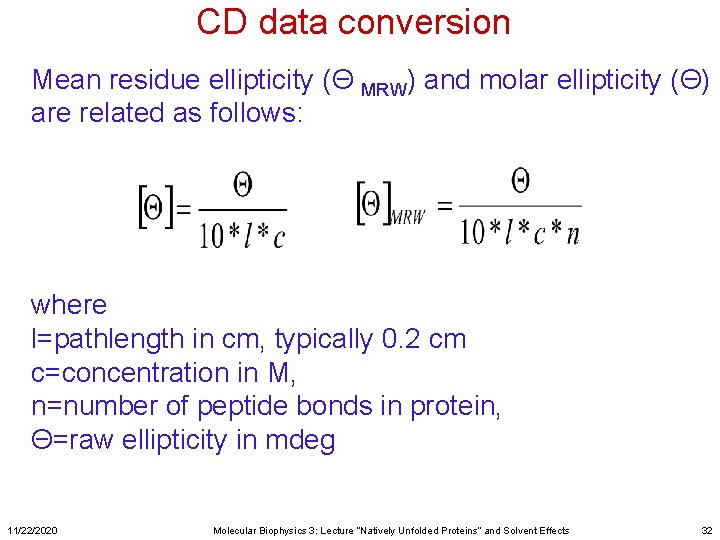
CD data conversion Mean residue ellipticity (Θ MRW) and molar ellipticity (Θ) are related as follows: where l=pathlength in cm, typically 0. 2 cm c=concentration in M, n=number of peptide bonds in protein, Θ=raw ellipticity in mdeg 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 32

Homework: CD Analysis Step 2 • do the prediction: e. g. Continll. exe • Prot. SS. out is the output file • Calc. CD. out allows you to check predicted versus observed CD spectra 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 33

Datafiles for Homework • There are two sets of data, a titration in trifluoroethanol and one in ammonium sulfate. – PBS. txt (no TFE, no AS) – 10 TFE. txt, 20 TFE. txt, 30 TFE. txt, 50 TFE. txt, 90 TFE. txt – 10 as. txt, 20 as. txt, 30 as. txt, 50 as. txt, 90 as. txt • Reminder: The data is raw ellipticity data, it needs to be converted: [Q]mrw = [Q]/(10*l*c*n) where [Q] is the raw ellipticity, l is the cell pathlength in cm, c is the protein concentration in M, n is the number of peptide bonds in the protein. This data is from p. Htr. II-cyt (28800 Da) that was measured at 0. 1 mg/ml in a 0. 2 cm cell. 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 34

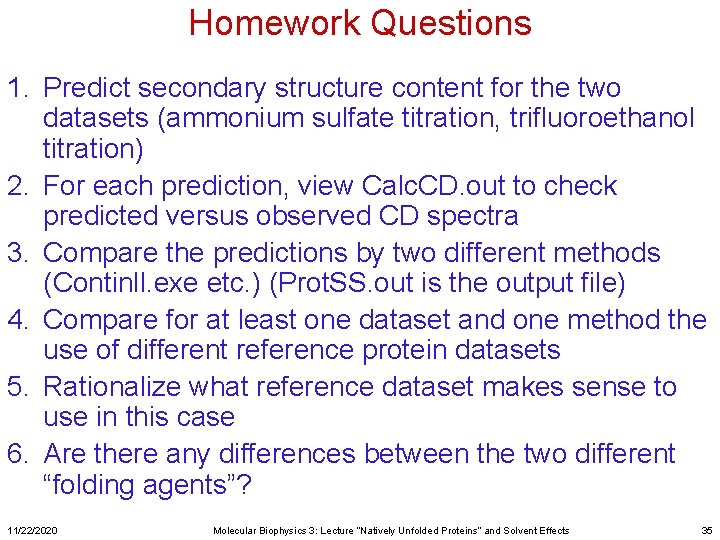
Homework Questions 1. Predict secondary structure content for the two datasets (ammonium sulfate titration, trifluoroethanol titration) 2. For each prediction, view Calc. CD. out to check predicted versus observed CD spectra 3. Compare the predictions by two different methods (Continll. exe etc. ) (Prot. SS. out is the output file) 4. Compare for at least one dataset and one method the use of different reference protein datasets 5. Rationalize what reference dataset makes sense to use in this case 6. Are there any differences between the two different “folding agents”? 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 35

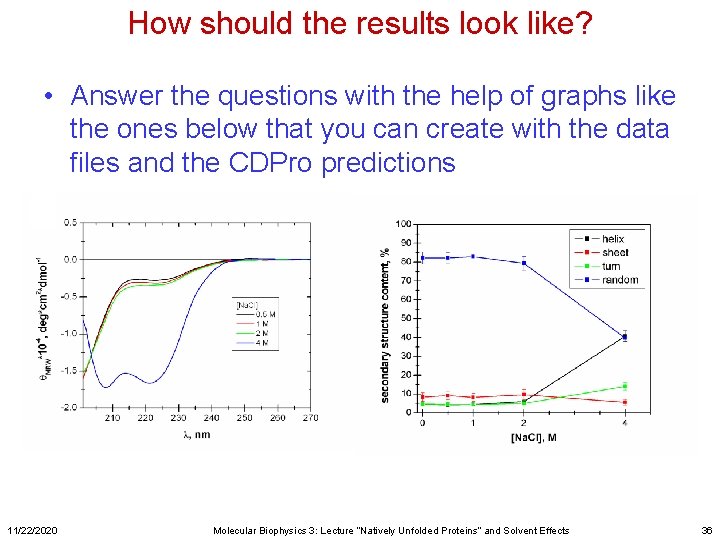
How should the results look like? • Answer the questions with the help of graphs like the ones below that you can create with the data files and the CDPro predictions 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 36

Lecture Overview • Natively unfolded proteins • Brief circular dichroism tutorial • Example CD: Is the transducer natively unfolded? • Brief intro to methods for “seeing” molecules • Example SANS: Is the transducer natively unfolded? • Theories on solvent effects • Alternative method for “seeing” molecules: AFM 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 37

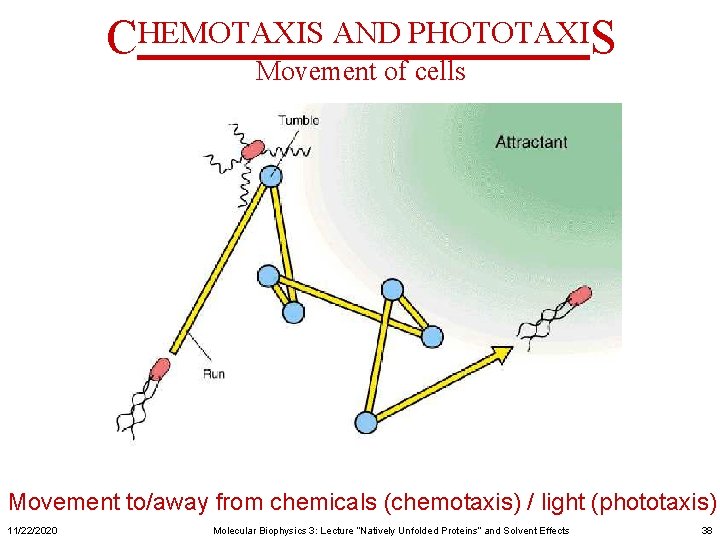
CHEMOTAXIS AND PHOTOTAXIS Movement of cells Movement to/away from chemicals (chemotaxis) / light (phototaxis) 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 38

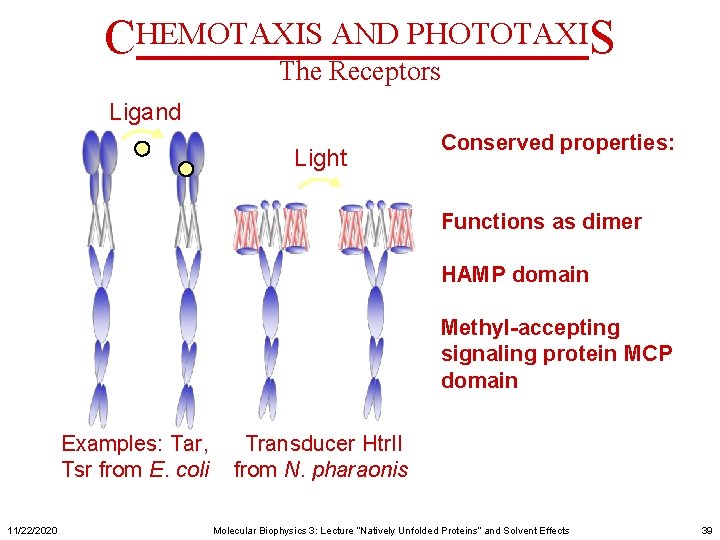
CHEMOTAXIS AND PHOTOTAXIS The Receptors Ligand Light Conserved properties: Functions as dimer HAMP domain Methyl-accepting signaling protein MCP domain Examples: Tar, Tsr from E. coli 11/22/2020 Transducer Htr. II from N. pharaonis Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 39

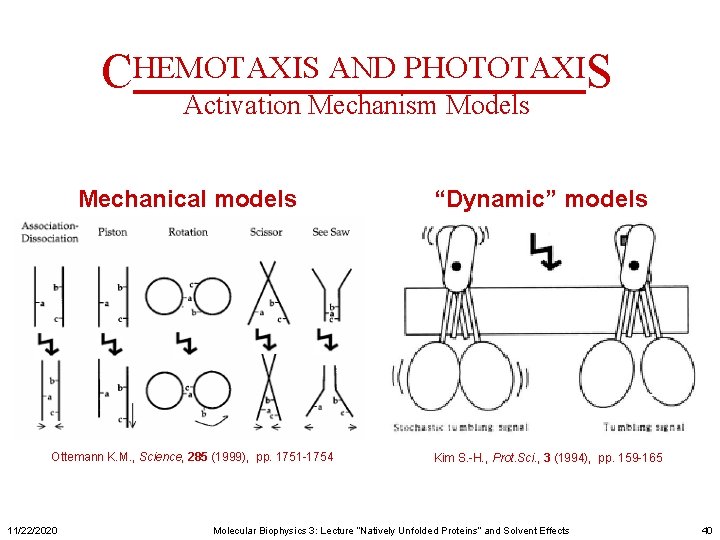
CHEMOTAXIS AND PHOTOTAXIS Activation Mechanism Models Mechanical models Ottemann K. M. , Science, 285 (1999), pp. 1751 -1754 11/22/2020 “Dynamic” models Kim S. -H. , Prot. Sci. , 3 (1994), pp. 159 -165 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 40

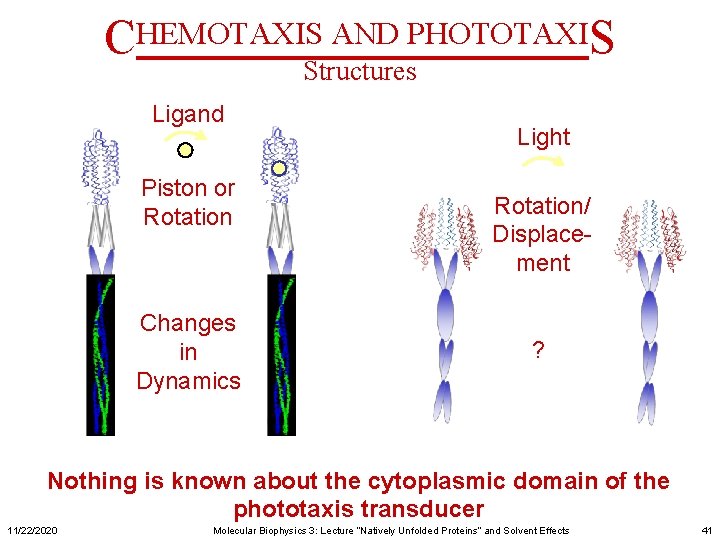
CHEMOTAXIS AND PHOTOTAXIS Structures Ligand Piston or Rotation Changes in Dynamics Light Rotation/ Displacement ? Nothing is known about the cytoplasmic domain of the phototaxis transducer 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 41

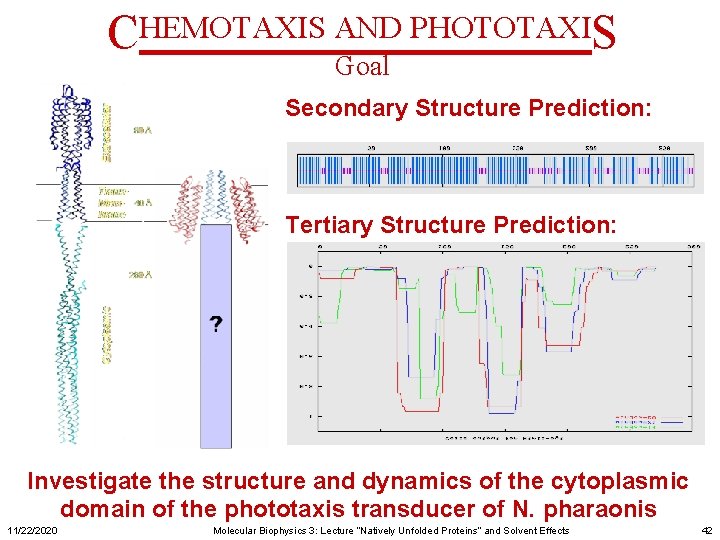
CHEMOTAXIS AND PHOTOTAXIS Goal Secondary Structure Prediction: Tertiary Structure Prediction: Investigate the structure and dynamics of the cytoplasmic domain of the phototaxis transducer of N. pharaonis 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 42

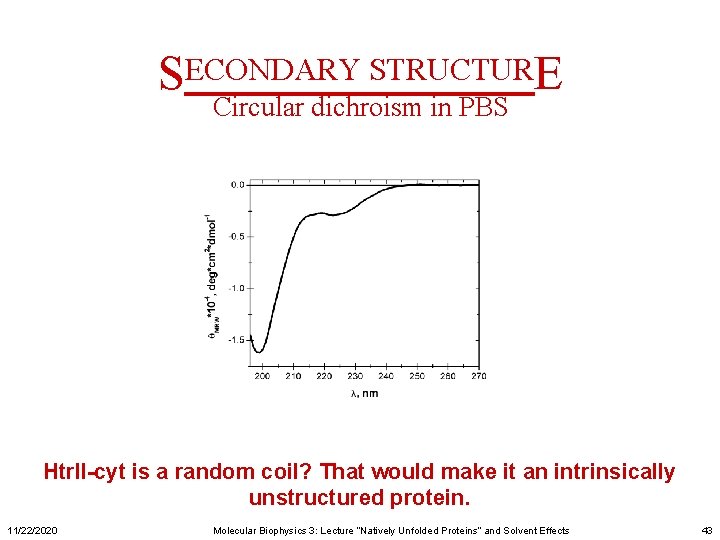
SECONDARY STRUCTURE Circular dichroism in PBS Htr. II-cyt is a random coil? That would make it an intrinsically unstructured protein. 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 43

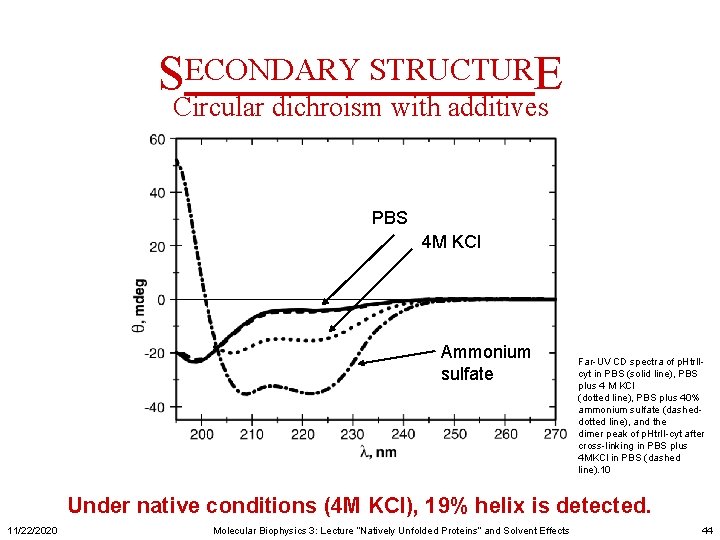
SECONDARY STRUCTURE Circular dichroism with additives PBS 4 M KCl Ammonium sulfate Far-UV CD spectra of p. Htr. IIcyt in PBS (solid line), PBS plus 4 M KCl (dotted line), PBS plus 40% ammonium sulfate (dasheddotted line), and the dimer peak of p. Htr. II-cyt after cross-linking in PBS plus 4 MKCl in PBS (dashed line). 10 Under native conditions (4 M KCl), 19% helix is detected. 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 44

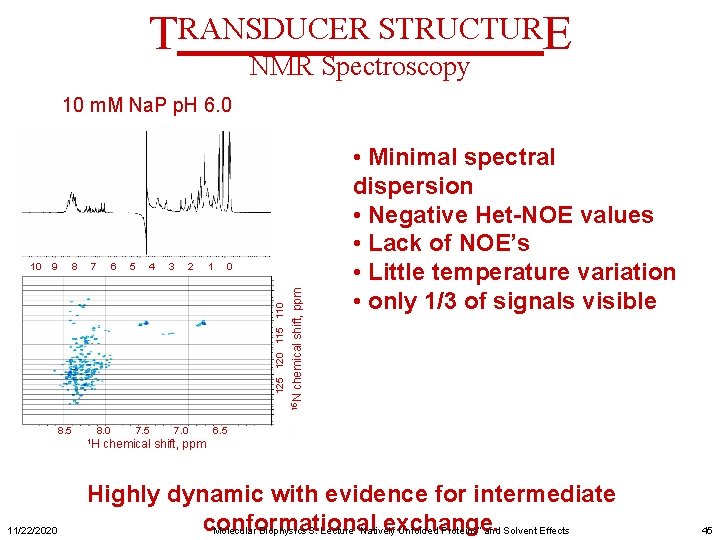
TRANSDUCER STRUCTURE NMR Spectroscopy 10 m. M Na. P p. H 6. 0 7 6 5 4 3 2 1 0 8. 5 8. 0 1 H 11/22/2020 7. 5 7. 0 chemical shift, ppm 8 15 N 9 125 120 115 110 10 • Minimal spectral dispersion • Negative Het-NOE values • Lack of NOE’s • Little temperature variation • only 1/3 of signals visible 6. 5 chemical shift, ppm Highly dynamic with evidence for intermediate conformational exchange. Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 45

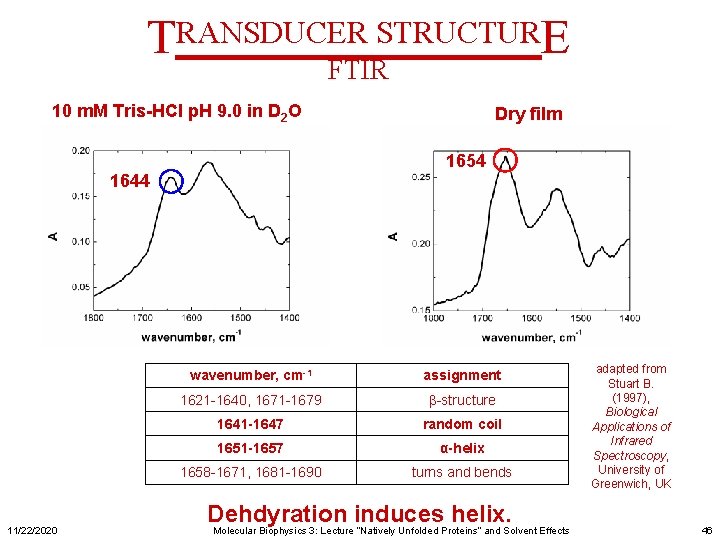
TRANSDUCER STRUCTURE FTIR 10 m. M Tris-HCl p. H 9. 0 in D 2 O 1654 1644 11/22/2020 Dry film wavenumber, cm-1 assignment 1621 -1640, 1671 -1679 β-structure 1641 -1647 random coil 1651 -1657 α-helix 1658 -1671, 1681 -1690 turns and bends Dehdyration induces helix. Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects adapted from Stuart B. (1997), Biological Applications of Infrared Spectroscopy, University of Greenwich, UK 46

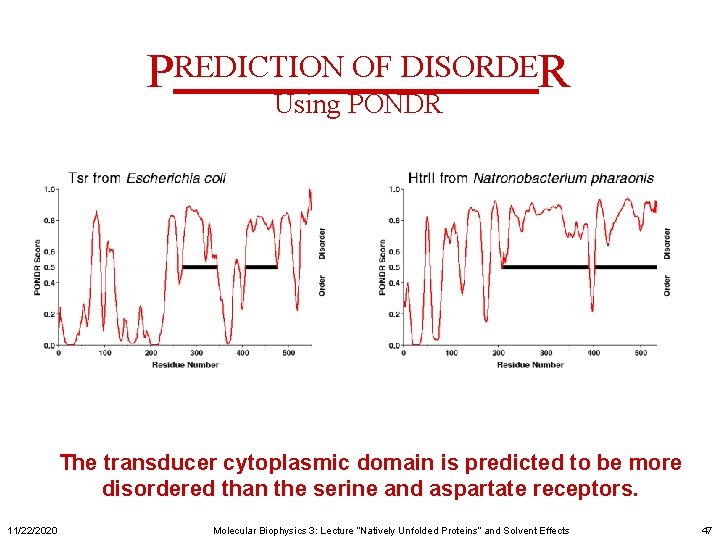
PREDICTION OF DISORDER Using PONDR The transducer cytoplasmic domain is predicted to be more disordered than the serine and aspartate receptors. 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 47

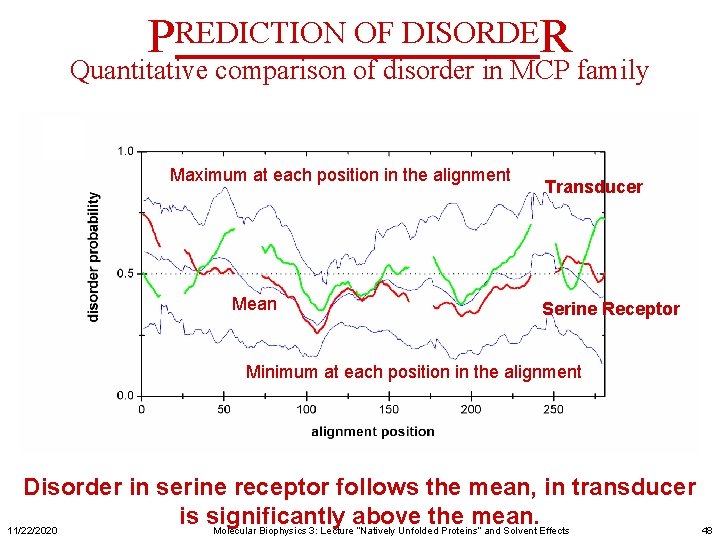
PREDICTION OF DISORDER Quantitative comparison of disorder in MCP family Maximum at each position in the alignment Mean Transducer Serine Receptor Minimum at each position in the alignment Disorder in serine receptor follows the mean, in transducer is significantly above the mean. 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 48

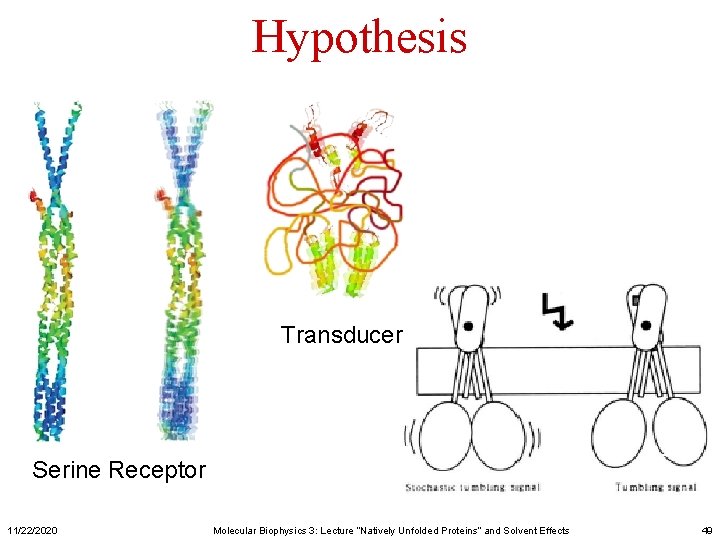
Hypothesis Transducer Serine Receptor 11/22/2020 Molecular Biophysics 3: Lecture “Natively Unfolded Proteins” and Solvent Effects 49
 Simbol dari weak entity
Simbol dari weak entity The great gatsby chapter 3 questions
The great gatsby chapter 3 questions Chapter 15 the last great nomadic challenges
Chapter 15 the last great nomadic challenges The last spin characters
The last spin characters Empowerment and participation in organizational behavior
Empowerment and participation in organizational behavior Pleasure and participation sports
Pleasure and participation sports Advantages of participation chart
Advantages of participation chart Spreading excellence and widening participation
Spreading excellence and widening participation Thank you so much for your attention
Thank you so much for your attention Thank you very much for your attention.
Thank you very much for your attention. Continuum of participation
Continuum of participation Vaict
Vaict Widening participation and spreading excellence
Widening participation and spreading excellence Financial literacy and stock market participation
Financial literacy and stock market participation Hình ảnh bộ gõ cơ thể búng tay
Hình ảnh bộ gõ cơ thể búng tay Bổ thể
Bổ thể Tỉ lệ cơ thể trẻ em
Tỉ lệ cơ thể trẻ em Voi kéo gỗ như thế nào
Voi kéo gỗ như thế nào Chụp phim tư thế worms-breton
Chụp phim tư thế worms-breton Chúa yêu trần thế alleluia
Chúa yêu trần thế alleluia Kể tên các môn thể thao
Kể tên các môn thể thao Thế nào là hệ số cao nhất
Thế nào là hệ số cao nhất Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới Cong thức tính động năng
Cong thức tính động năng Trời xanh đây là của chúng ta thể thơ
Trời xanh đây là của chúng ta thể thơ Mật thư anh em như thể tay chân
Mật thư anh em như thể tay chân Làm thế nào để 102-1=99
Làm thế nào để 102-1=99 Phản ứng thế ankan
Phản ứng thế ankan Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới Thể thơ truyền thống
Thể thơ truyền thống Quá trình desamine hóa có thể tạo ra
Quá trình desamine hóa có thể tạo ra Một số thể thơ truyền thống
Một số thể thơ truyền thống Bàn tay mà dây bẩn
Bàn tay mà dây bẩn Vẽ hình chiếu vuông góc của vật thể sau
Vẽ hình chiếu vuông góc của vật thể sau Thế nào là sự mỏi cơ
Thế nào là sự mỏi cơ đặc điểm cơ thể của người tối cổ
đặc điểm cơ thể của người tối cổ Thế nào là giọng cùng tên
Thế nào là giọng cùng tên Vẽ hình chiếu đứng bằng cạnh của vật thể
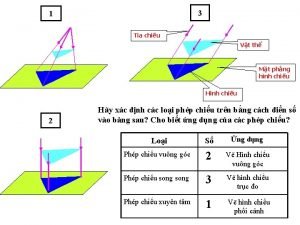
Vẽ hình chiếu đứng bằng cạnh của vật thể Tia chieu sa te
Tia chieu sa te Thẻ vin
Thẻ vin đại từ thay thế
đại từ thay thế điện thế nghỉ
điện thế nghỉ Tư thế ngồi viết
Tư thế ngồi viết Diễn thế sinh thái là
Diễn thế sinh thái là Dạng đột biến một nhiễm là
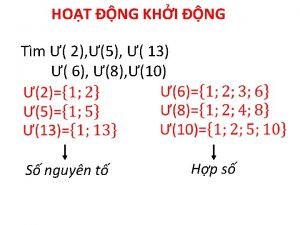
Dạng đột biến một nhiễm là So nguyen to
So nguyen to Tư thế ngồi viết
Tư thế ngồi viết Lời thề hippocrates
Lời thề hippocrates Thiếu nhi thế giới liên hoan
Thiếu nhi thế giới liên hoan