Graphics Plots matplotlib pylab BCHB 524 Lecture 24
Graphics & Plots: matplotlib & pylab BCHB 524 Lecture 24 BCHB 524 - Edwards

Outline l Testing pylab l Download data l Basic plots l l scatter plots, histograms, boxplots Exercises BCHB 524 - Edwards 2
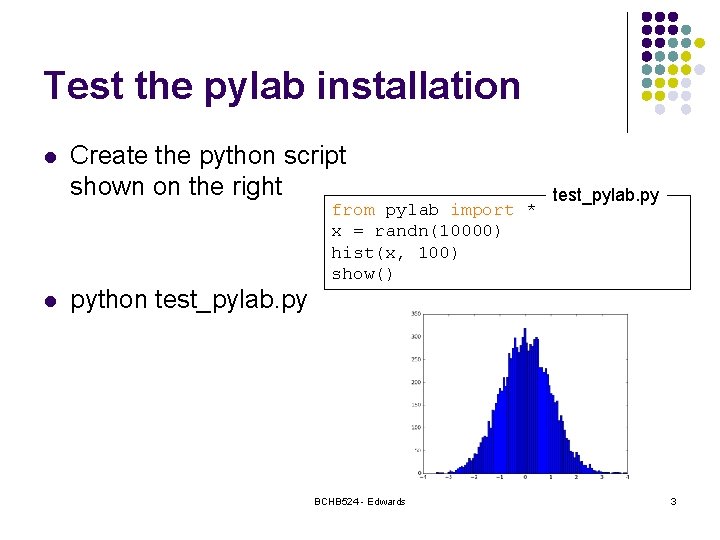
Test the pylab installation l l Create the python script shown on the right python test_pylab. py from pylab import * x = randn(10000) hist(x, 100) show() BCHB 524 - Edwards test_pylab. py 3

Download some data l Download the data and the module for handling it, from the course homepage l l data. txt, data. py Take a look! l l Open data. txt in a text-editor (IDLE or notepad) look. py Run look. py from data import * print genes print data['AA 055368'] print t 1 data['AA 055368'] BCHB 524 - Edwards 4
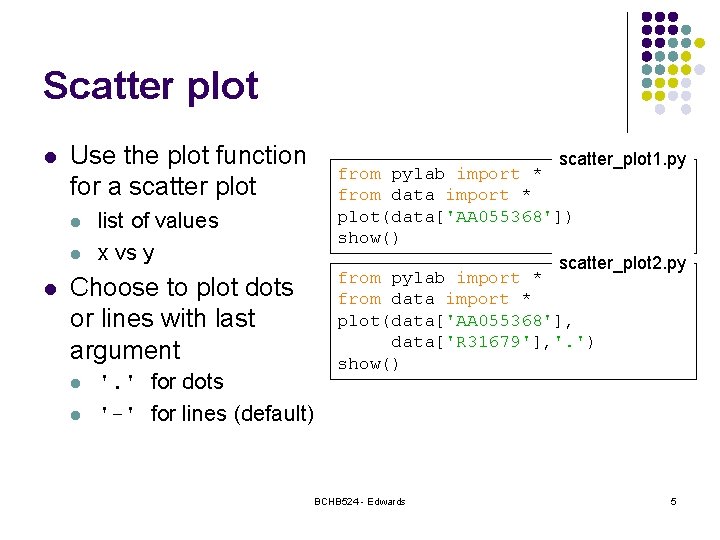
Scatter plot l Use the plot function for a scatter plot l list of values x vs y Choose to plot dots or lines with last argument l l '. ' for dots '-' for lines (default) scatter_plot 1. py from pylab import * from data import * plot(data['AA 055368']) show() scatter_plot 2. py from pylab import * from data import * plot(data['AA 055368'], data['R 31679'], '. ') show() BCHB 524 - Edwards 5

Heatmap l Use the pcolor function for a heatmap from pylab import * from data import * pcolor(tmdata) show() heatmap 1. py list of lists, or numpy 2 -D matrix from pylab import * heatmap 2. py from data import * l pcolor(tmdata) l Choose colormap clim((-6, 6)) gci(). set_cmap(cm. Rd. Yl. Gn) l cool() colorbar() l hot() ylim([nsmpl, 0]) axis('tight') l Lots of tweaking options xlabel('Gene') to make it ylabel('Sample') look just right show() # savefig('colormap. png', dpi=150) l BCHB 524 - Edwards 6
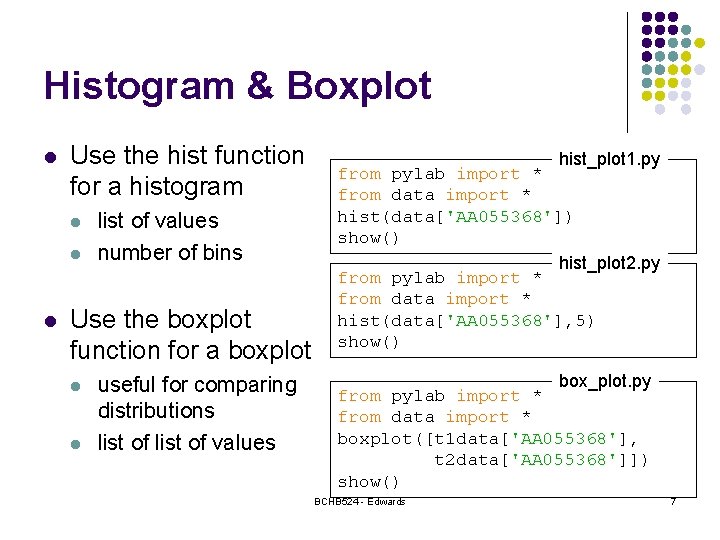
Histogram & Boxplot l Use the hist function for a histogram l list of values number of bins Use the boxplot function for a boxplot l l useful for comparing distributions list of values hist_plot 1. py from pylab import * from data import * hist(data['AA 055368']) show() hist_plot 2. py from pylab import * from data import * hist(data['AA 055368'], 5) show() box_plot. py from pylab import * from data import * boxplot([t 1 data['AA 055368'], t 2 data['AA 055368']]) show() BCHB 524 - Edwards 7
Check out the matplotlib gallery! BCHB 524 - Edwards 8
Lets analyze this dataset! l Find differentially expressed genes! differential. py from pylab import * from data import * g 2 t = {} for g in genes: g 2 t[g] = tstatistic(t 1 data[g], t 2 data[g]) x = g 2 t. values() hist(x) show() bytstat = sorted(genes, key=g 2 t. get) print "Min: ", bytstat[0], min(x) print "Max: ", bytstat[-1], max(x) BCHB 524 - Edwards 9

Lets analyze this dataset! l Find differentially expressed genes! differential 1. py from pylab import * from data import * g 2 t = {} for g in genes: g 2 t[g] = tstatistic(t 1 data[g], t 2 data[g]) bytstat = sorted(genes, key=g 2 t. get) gene = bytstat[0] boxplot([t 1 data[gene], t 2 data[gene]]) title(gene) show() BCHB 524 - Edwards 10

Find correlated genes correlated. py from pylab import * from data import * gp 2 rho = {} for i in range(ngene): for j in range(i+1, ngene): gi = genes[i] gj = genes[j] gp 2 rho[(gi, gj)] = corrcoef(data[gi], data[gj])[0, 1] hist(gp 2 rho. values()) show() sx = sorted(gp 2 rho. keys(), key=gp 2 rho. get) print sx[0], sx[-1] BCHB 524 - Edwards 11

Find correlated genes correlated 1. py from pylab import * from data import * gp 2 rho = {} for i in range(ngene): for j in range(i+1, ngene): gi = genes[i] gj = genes[j] gp 2 rho[(gi, gj)] = corrcoef(data[gi], data[gj])[0, 1] sx = sorted(gp 2 rho. keys(), key=gp 2 rho. get) bestpair = sx[-1] gi = bestpair[0] gj = bestpair[1] plot(data[gi], data[gj], '. ') show() BCHB 524 - Edwards 12

Exercises l Try each of the examples shown in these slides. l Check out the gallery of figures on the matplotlib web-site. l Write a program to plot the GC % of 20 -mer DNA windows from a DNA sequence. BCHB 524 - Edwards 13
- Slides: 13