GO the Gene Ontology Functional enrichment analysis F









- Slides: 24

GO : the Gene Ontology & Functional enrichment analysis F. Burdet

Brief presentation of GO Slides about GO used with the kind permission of: Amelia Ireland GO Curator EBI, Cambridge, UK

What is the Gene Ontology? • Definition: w The Gene Ontology: “a controlled vocabulary that can be applied to all organisms even as knowledge of gene and protein roles in cells is accumulating and changing” • A controlled vocabulary to describe gene products - proteins and RNA - in any organism.

What is GO? • One of the Open Biological Ontologies • Standard, species-neutral way of representing biology • Three structured networks of defined terms to describe gene product attributes • More like a phrase book than a biology text book

How does GO work? What information might we want to capture about a gene product? • What does the gene product do? • Where and when does it act? • Why does it perform these activities?

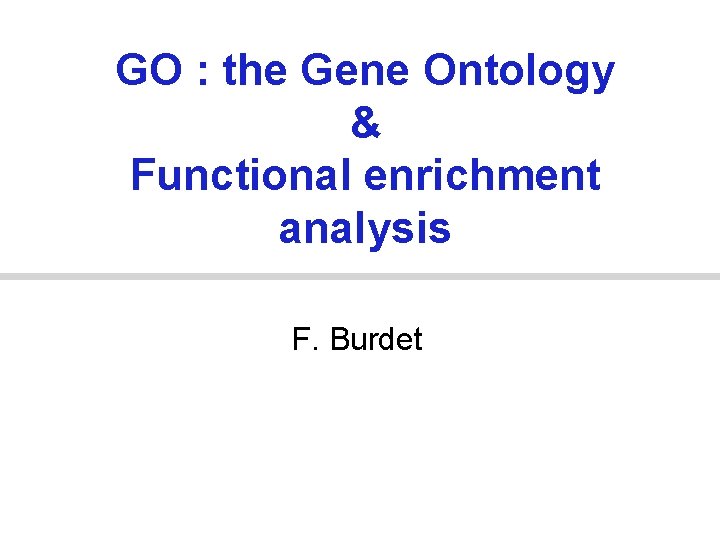
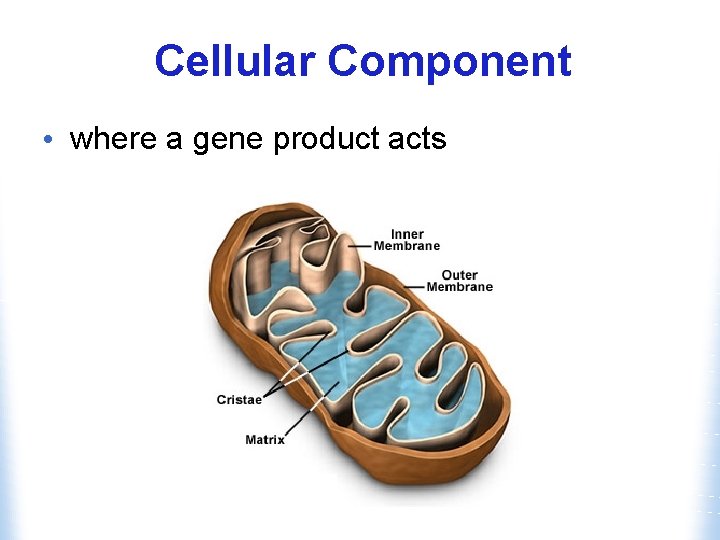
Cellular Component • where a gene product acts

Cellular Component

Cellular Component

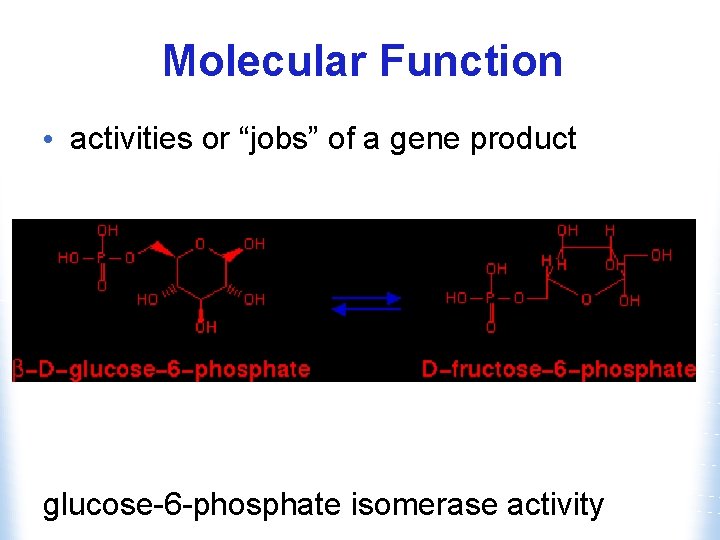
Molecular Function • activities or “jobs” of a gene product glucose-6 -phosphate isomerase activity

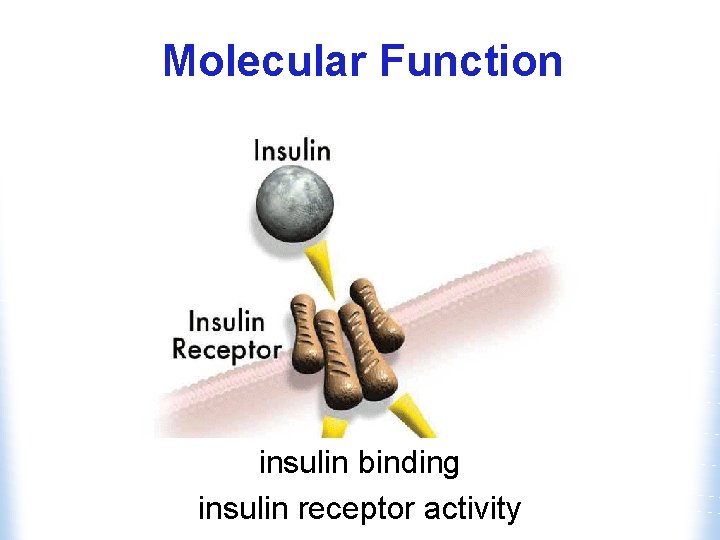
Molecular Function insulin binding insulin receptor activity

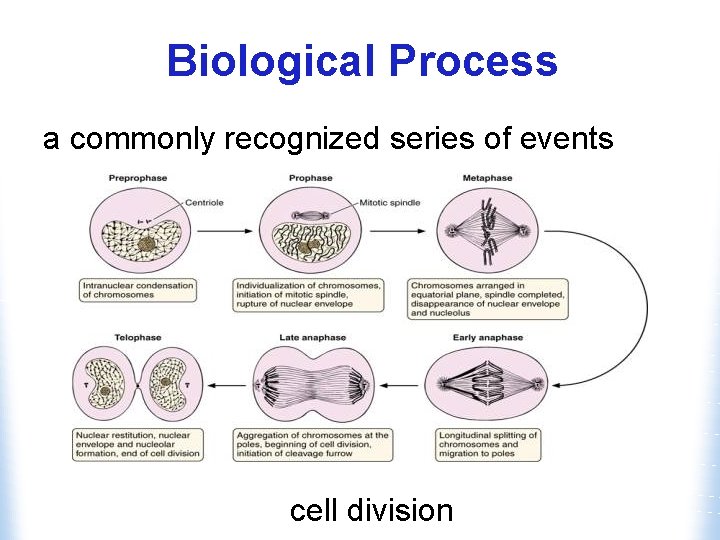
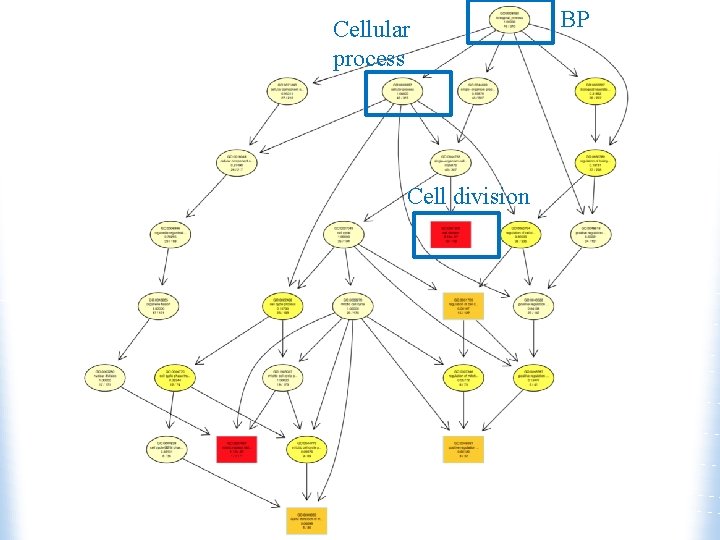
Biological Process a commonly recognized series of events cell division

Biological Process transcription

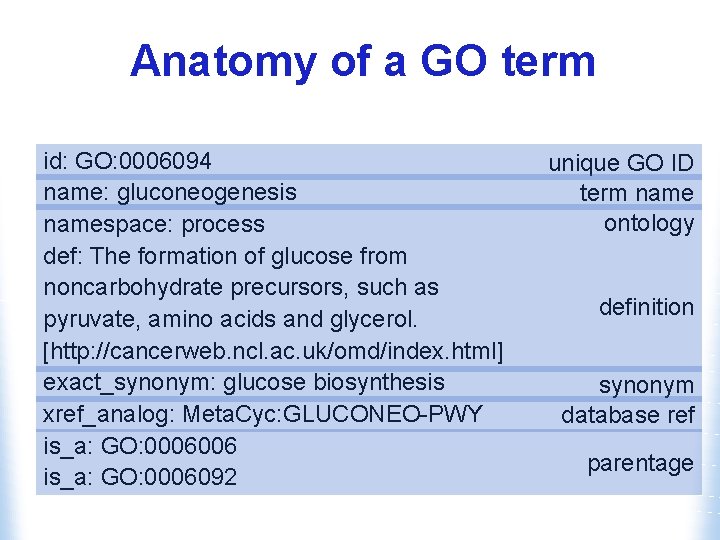
Anatomy of a GO term id: GO: 0006094 name: gluconeogenesis namespace: process def: The formation of glucose from noncarbohydrate precursors, such as pyruvate, amino acids and glycerol. [http: //cancerweb. ncl. ac. uk/omd/index. html] exact_synonym: glucose biosynthesis xref_analog: Meta. Cyc: GLUCONEO-PWY is_a: GO: 0006006 is_a: GO: 0006092 unique GO ID term name ontology definition synonym database ref parentage

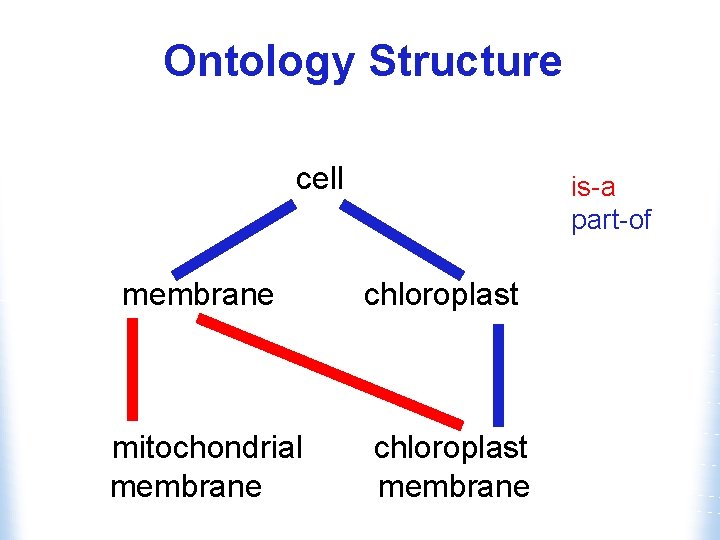
Ontology Structure • Ontologies are structured as a hierarchical directed acyclic graph • Terms can have more than one parent and zero, one or more children • Terms are linked by two relationships w is-a w part-of

Ontology Structure cell membrane mitochondrial membrane is-a part-of chloroplast membrane

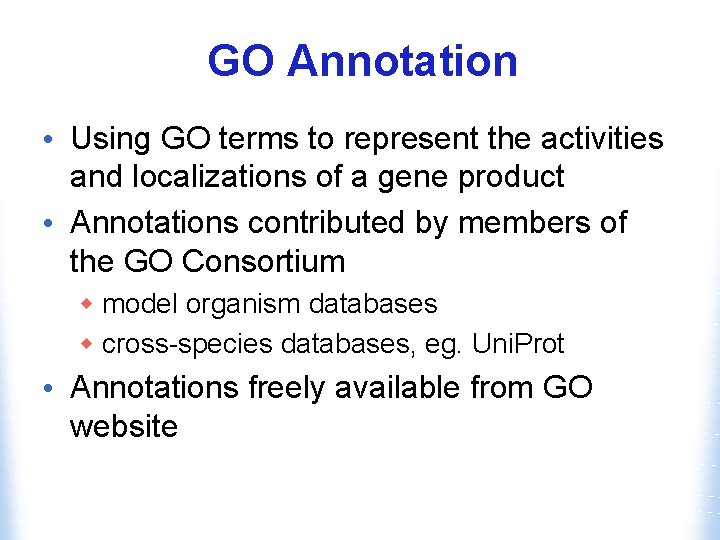
GO Annotation • Using GO terms to represent the activities and localizations of a gene product • Annotations contributed by members of the GO Consortium w model organism databases w cross-species databases, eg. Uni. Prot • Annotations freely available from GO website

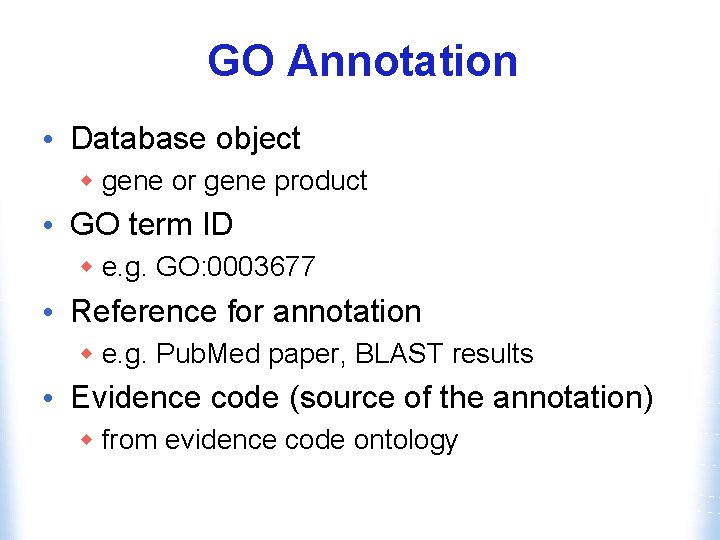
GO Annotation • Database object w gene or gene product • GO term ID w e. g. GO: 0003677 • Reference for annotation w e. g. Pub. Med paper, BLAST results • Evidence code (source of the annotation) w from evidence code ontology

Functional enrichment analysis

An other way to look at the gene modules • I have a set of genes (from a module). On top of the correlations with the traits, can I discover more about it? • One way is to look if some known pathways or functionally related genes are enriched in this module • How can one assess the enrichment?

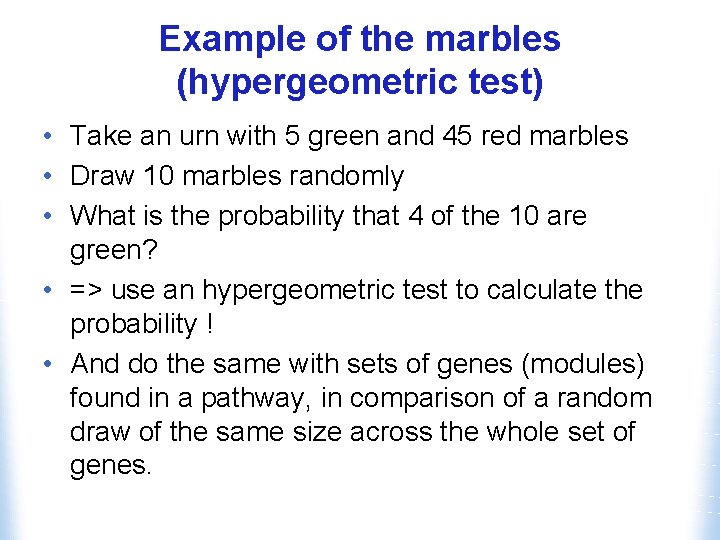
Example of the marbles (hypergeometric test) • Take an urn with 5 green and 45 red marbles • Draw 10 marbles randomly • What is the probability that 4 of the 10 are green? • => use an hypergeometric test to calculate the probability ! • And do the same with sets of genes (modules) found in a pathway, in comparison of a random draw of the same size across the whole set of genes.

From marbles to genes • Say I have 100 genes in my orange module • 25 out of the 30 genes involved in lipid biosynthesis are found in my orange module • Given the fact that there are 10’ 000 genes in the dataset… • What was the probability of this distribution to happen, compared to a random draw?

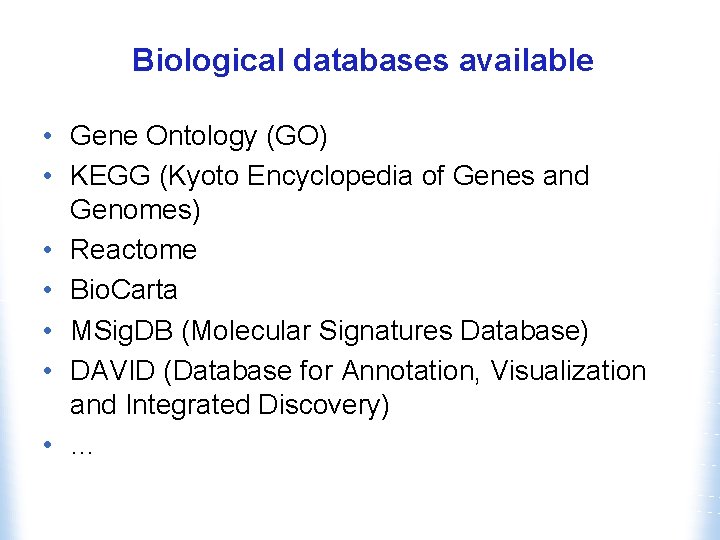
Biological databases available • Gene Ontology (GO) • KEGG (Kyoto Encyclopedia of Genes and Genomes) • Reactome • Bio. Carta • MSig. DB (Molecular Signatures Database) • DAVID (Database for Annotation, Visualization and Integrated Discovery) • …

top. GO • Bioconductor package • Has an algorithm that will only consider the significant “leaves” instead of all the levels (weight)

Cellular process Cell division BP