GO Software Group 2006 Regulation terms Regulation DAGs



![[Term] id: GO: 0050855 name: regulation of B cell receptor signaling pathway namespace: biological_process [Term] id: GO: 0050855 name: regulation of B cell receptor signaling pathway namespace: biological_process](https://slidetodoc.com/presentation_image/b727829671dc680e1e32e9d67471156e/image-7.jpg)










- Slides: 21

GO Software Group 2006

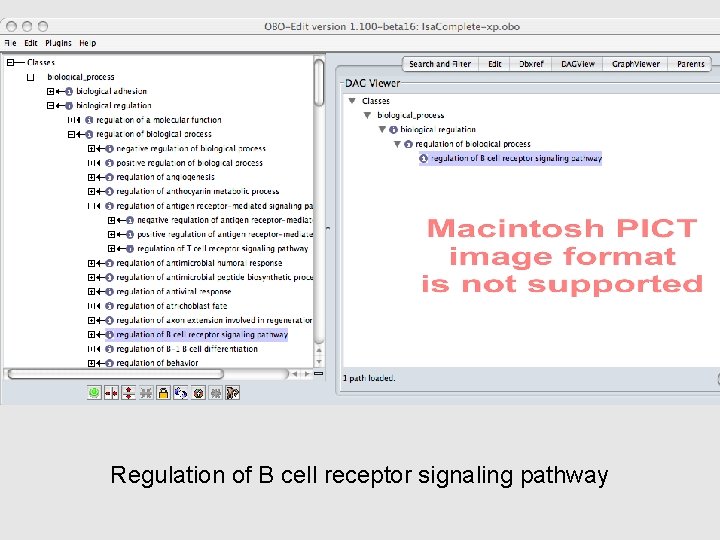
Regulation terms • Regulation DAGs are COMPLEX and TANGLED!

Regulation of B cell receptor signaling pathway

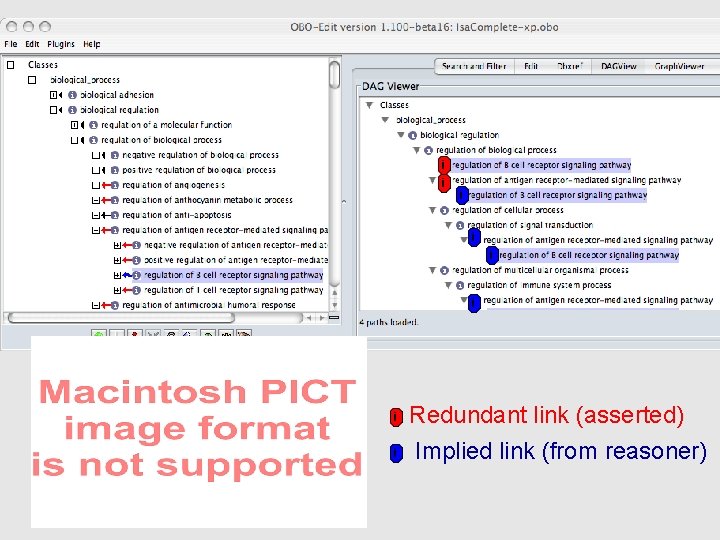
i i i i Redundant link (asserted) Implied link (from reasoner)

This entire DAG can be computed automatically

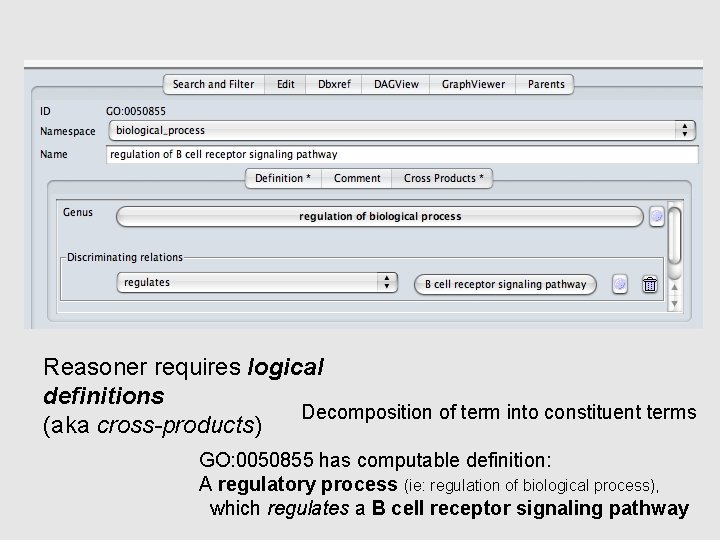
How does the oboedit reasoner do this? ? Reasoner requires logical definitions Decomposition of term into constituent terms (aka cross-products) GO: 0050855 has computable definition: A regulatory process (ie: regulation of biological process), which regulates a B cell receptor signaling pathway
![Term id GO 0050855 name regulation of B cell receptor signaling pathway namespace biologicalprocess [Term] id: GO: 0050855 name: regulation of B cell receptor signaling pathway namespace: biological_process](https://slidetodoc.com/presentation_image/b727829671dc680e1e32e9d67471156e/image-7.jpg)
[Term] id: GO: 0050855 name: regulation of B cell receptor signaling pathway namespace: biological_process def: "Any process that modulates the frequency, rate or extent of signaling pathways initiated by the cross-linking of an antigen receptor on a B cell. " [GOC: ai] is_a: GO: 0050789 ! regulation of biological process relationship: regulates GO: 0050853 ! B cell receptor signaling pathway intersection_of: GO: 0050789 ! regulation of biological process intersection_of: regulates GO: 0050853 ! B cell receptor signaling pathway

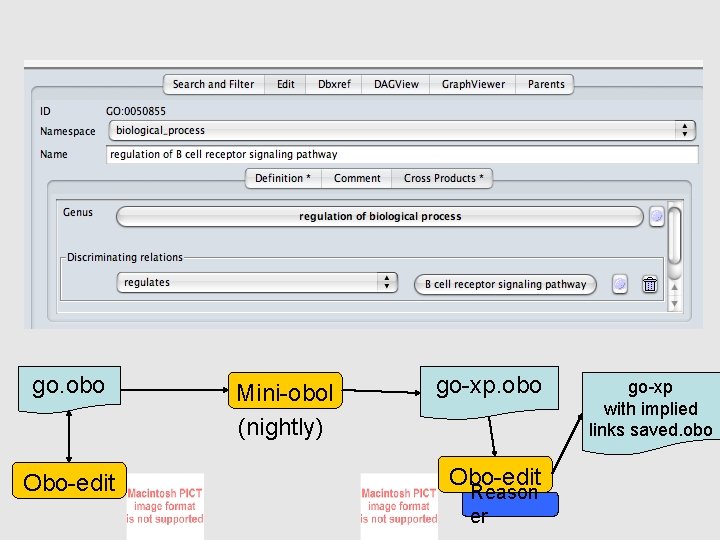
go. obo Obo-edit Mini-obol (nightly) go-xp. obo Obo-edit Reason er go-xp with implied links saved. obo

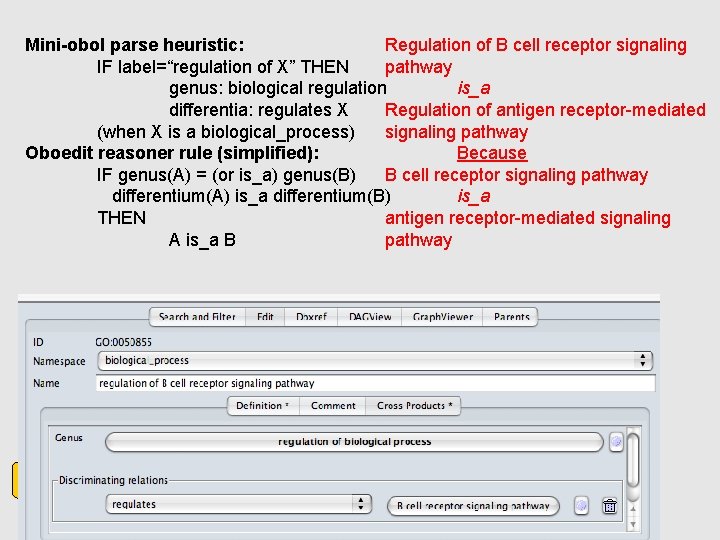
Mini-obol parse heuristic: Regulation of B cell receptor signaling IF label=“regulation of X” THEN pathway genus: biological regulation is_a differentia: regulates X Regulation of antigen receptor-mediated (when X is a biological_process) signaling pathway Oboedit reasoner rule (simplified): Because IF genus(A) = (or is_a) genus(B) B cell receptor signaling pathway differentium(A) is_a differentium(B) is_a THEN antigen receptor-mediated signaling A is_a B pathway go. obo Obo-edit Mini-obol (nightly) go-xp. obo Obo-edit Reason er


B cell receptor signaling pathway

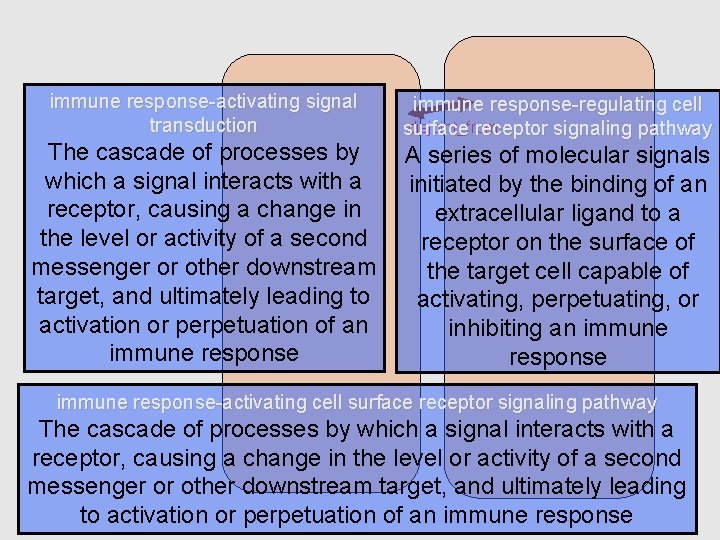
immune response-activating signal transduction The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately leading to activation or perpetuation of an immune response-regulating cell disjoint_from surface receptor signaling pathway A series of molecular signals initiated by the binding of an extracellular ligand to a receptor on the surface of the target cell capable of activating, perpetuating, or inhibiting an immune response-activating cell surface receptor signaling pathway The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately leading to activation or perpetuation of an immune response

Next steps: regulation terms • Add logical definitions directly to go_edit. obo – Remove dependency on heuristic obol term parsing – Curators actively maintain logical definitions • Win: – Maintenance of regulation DAG automated • Potential problems: – Reasoner slows oboedit down – Actively being addressed

Logical definitions in other parts of GO • Sequence Ontology – 156 Logical definitions – Automates a lot of DAG maintenance • GO terms that refer to types of cell – In progress • Go live with regulation cross-products first • Others – – Regulation of qualities (PATO) CC x CC Chemical Anatomy


SWUG Progress Highlights 2006 • Applications – Amigo 1 – Amigo 2 – OBO-Edit • Database and perl tools – – Subsets/slims GO. xrf_abs Go-db-perl released to CPAN Term enrichment added to API • Ontology development support – Obo-format 1. 2 – Logical definitions (regulation, cell, SO)

SO x GO • What kinds of entities to SO terms represent? – Molecules? – Physical sequences? – Abstract sequences?

Pre- and post- composition in GO • Pre-composition – Terms such as oocyte growth are present with IDs in GO – Ideally the term is decomposed into a genusdifferentia (cross-product) definition which is also stored in gene_ontology_edit. obo • Post-composition – Terms such as cholesterol transport in the liver are dynamically composed at annotation time – Composed term need not be in GO • But constituent terms (cholesterol transport, liver) should be in GO or other ontologies

Implementing postcomposition • The two approaches are complementary • We should extend the annotation format to allow denoting more specific classes – e. g. • cholesterol transport in liver – advanced applications can query this – standard applications suffer no loss – extended annotations can be used to help seed new terms in the ontology • This is already being done (MGI, Dicty) – we just want to capture this in interopeable way

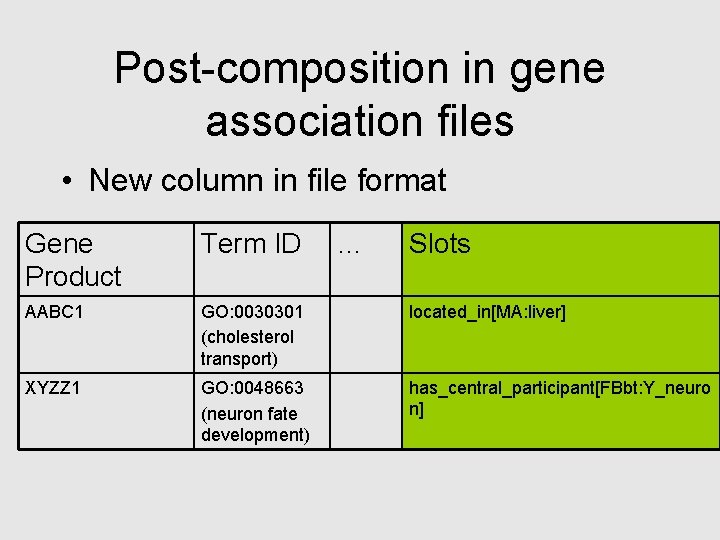
Post-composition in gene association files • New column in file format Gene Product Term ID … Slots AABC 1 GO: 0030301 (cholesterol transport) located_in[MA: liver] XYZZ 1 GO: 0048663 (neuron fate development) has_central_participant[FBbt: Y_neuro n]

Important note on postcomposition • This is not an either-or situation • We will retain pre-composed terms – terms will continue to be created for real biological types • Annotation post-composition can be used to further refine existing pre-composed terms – if the post-composed term is later created in the GO, the annotation can be automatically migrated • Tools can ignore post-composition for small loss in specificity – defaults to the current paradigm