GMS 6644 Apoptosis Lecture 5 p 53 and







- Slides: 44

GMS 6644 Apoptosis Lecture 5: p 53 and Apoptosis R 4 -265 Mar 1, 06, 9 – 10: 30 pm Dr. Daiqing Liao B 1 -016 dliao@ufl. edu

Tumor-suppressor genes, function like brakes, keep cell numbers down, either by inhibiting progress through the cell cycle and thereby preventing cell birth, or by promoting programmed cell death (also called apoptosis). When cellular tumor suppressor genes are rendered non-functional through mutation, the cell becomes malignant. Examples are the gene encoding the retinoblastoma protein (Rb), inactivated in retinoblastomas, p 53, and p 16 INK 4 a, which inhibits cyclin-dependent kinases and is inactivated in many different tumors.

Oncogenes stimulate appropriate cell growth under normal conditions, as required for the continued turnover and replenishment of the skin, gastrointestinal tract and blood, for example. Cells with mutant oncogenes continue to grow (or refuse to die) even when they are receiving no growth signals. Examples are Ras, activated in pancreatic and colon cancers, and Bcl-2, activated in lymphoid tumours. Amplification of oncogenes (more than their normal gene copy number) is also found in cancer: MDM 2 is amplified in liposarcomas.

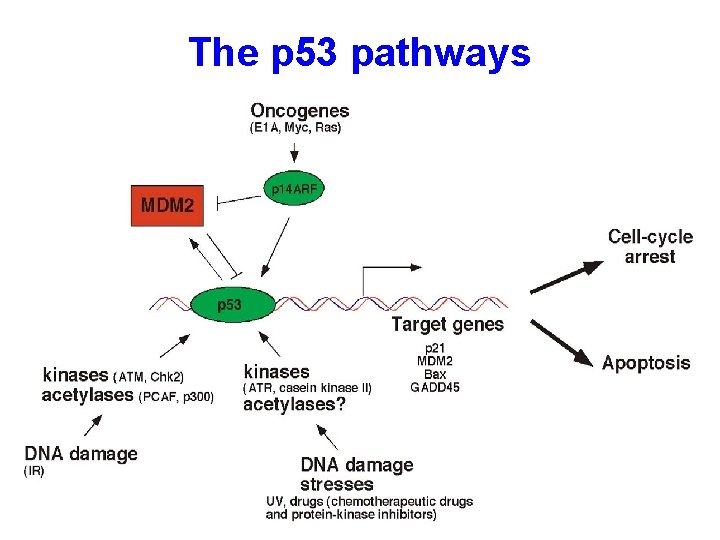
Roles of p 53 in apoptosis 1. p 53 induces apoptosis through transcriptional activation of proapoptotic genes, such as Puma, Noxa, p 53 AIP 1, Bax, Apaf-1 etc. 2. It can also directly induce apoptosis by localizing to mitochondria via interaction with Bcl-2 family protein Bcl-x. L and facilitating Bax oligomerization Reading: Vousden and Lu: Nature Reviews Cancer, 2002, 2: 594 -604.

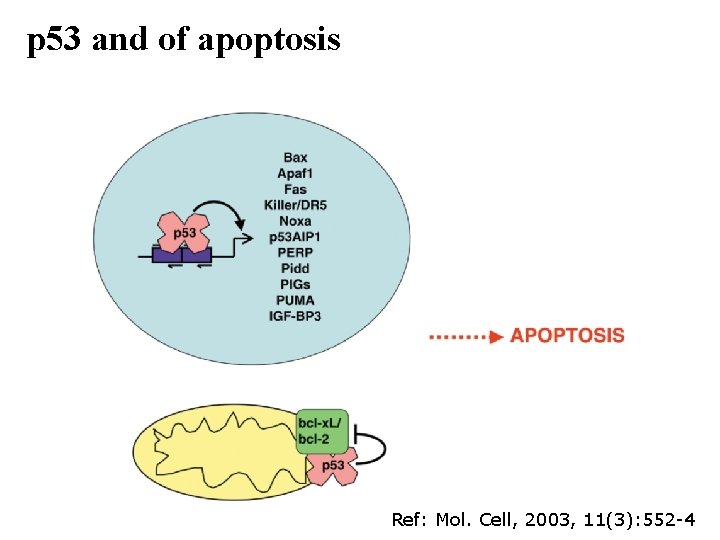
p 53 and of apoptosis Ref: Mol. Cell, 2003, 11(3): 552 -4

p 53 and apoptosis Ref: Cell, 2002, 108: 153 -164

Tumor Suppressor p 53 • First identified as a protein associated with viral oncogenes • Mutated/inactivated in a majority of human cancers • Integrates numerous signals that control cell life and death • A common denominator in human cancer • Understanding functions and regulation of p 53 is of great importance in cancer biology and cancer therapy


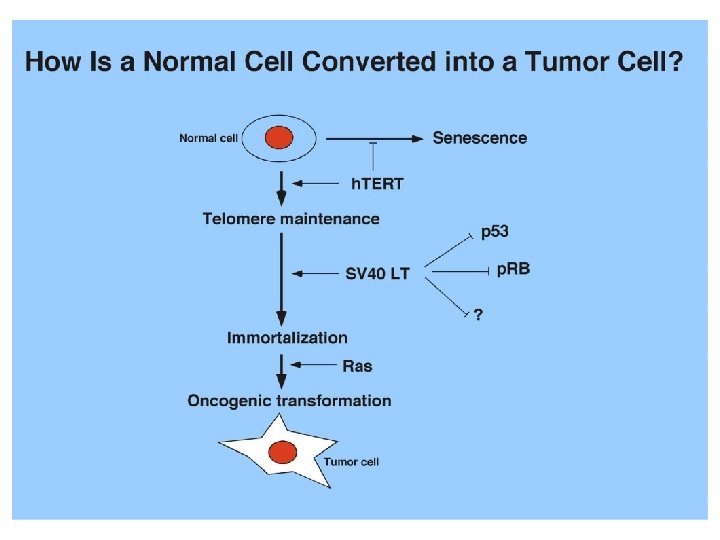
The p 53 pathways

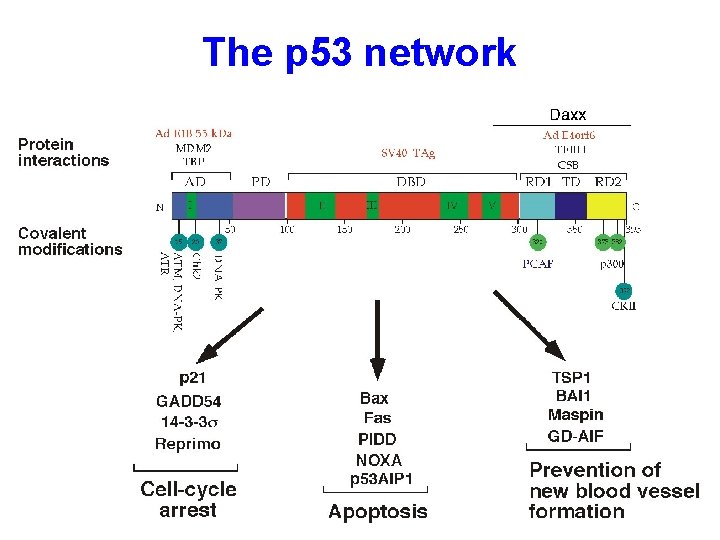
The p 53 network

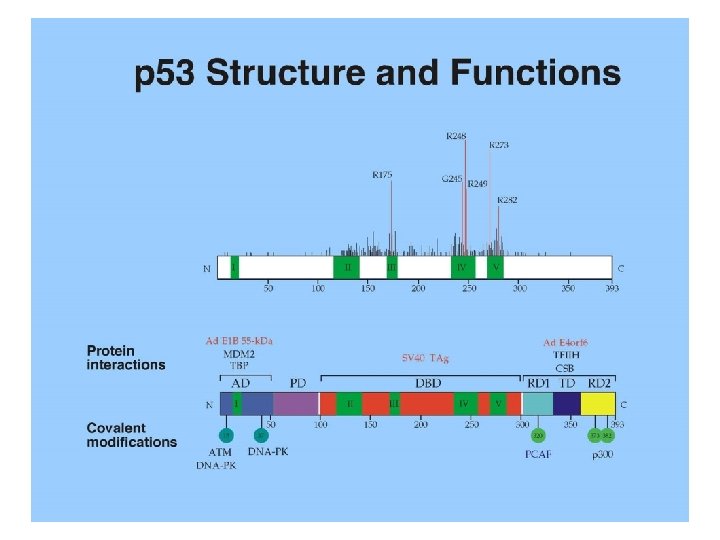
p 53 is a sequence-specific DNA binding protein 1. p 53 central core-domain interacts directly with DNA 2. p 53 binding sites consist of four copies of the pentamer consensus sequence Pu. Pu. C(A/T). The pentamers are oriented in alternating directions. A short stretch of sequence up to 13 bp may be inserted between the pairs of pentamers. The p 53 target genes in the human genome usually carry the consensus sequence. 3. Amino acid residues in the core-domain that are critical for DNA-binding are among the “hot-spots” of tumorderived p 53 mutations, attesting to the importance of DNA-binding for p 53’s tumor suppression function.

Structure of p 53 core-domain Ref: Science, 265: 346 -355, 1994

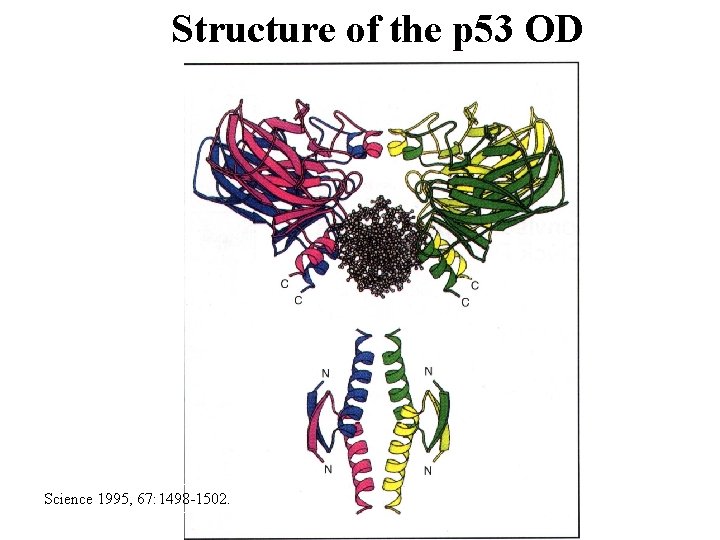
Structure of the p 53 OD Science 1995, 67: 1498 -1502.

The p 53 -MDM 2 feedback loop 1. MDM 2 binds to p 53 N-terminal transactivation domain and inhibits p 53 -dependent transcription. 2. 2. MDM 2 is a transcription target of p 53. 3. MDM 2 is an E 3 ubiquitin ligase of p 53, thus targeting p 53 for proteolytic degradation. 4. MDM 2 knockout is lethal for mouse embryonic development, but simultaneous deletion of p 53 and MDM 2 genes rescues MDM 2 -KO, thus confirming the in vivo genetic interaction of these two proteins.

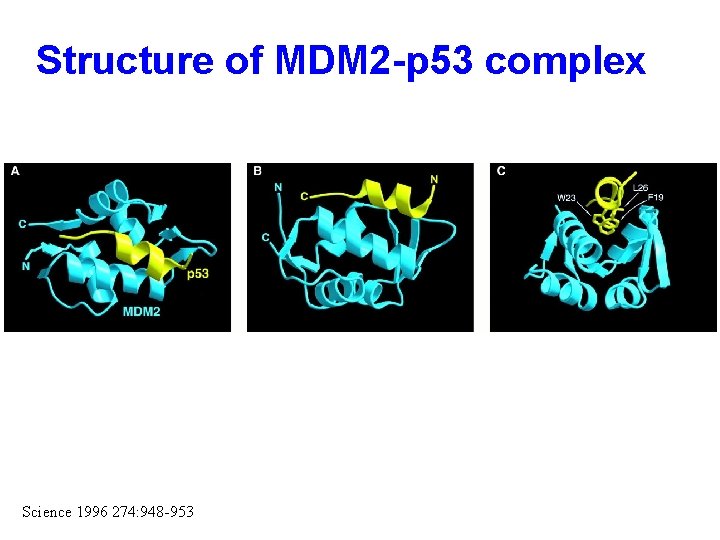
Structure of MDM 2 -p 53 complex Science 1996 274: 948 -953

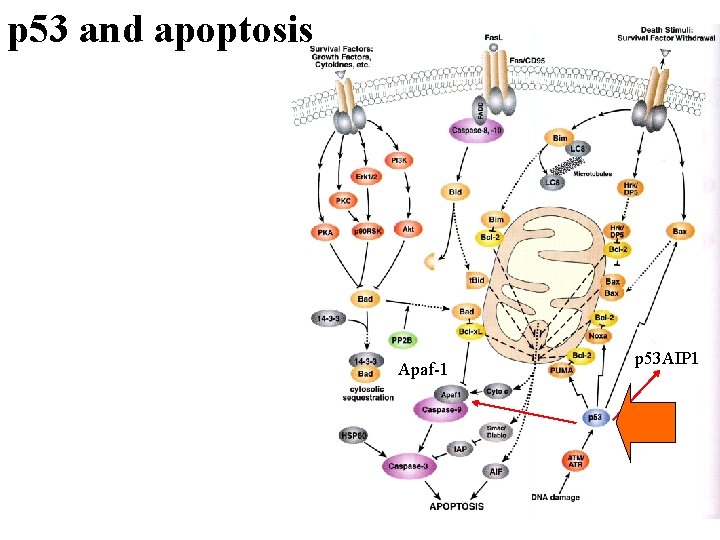
p 53 and apoptosis Apaf-1 p 53 AIP 1

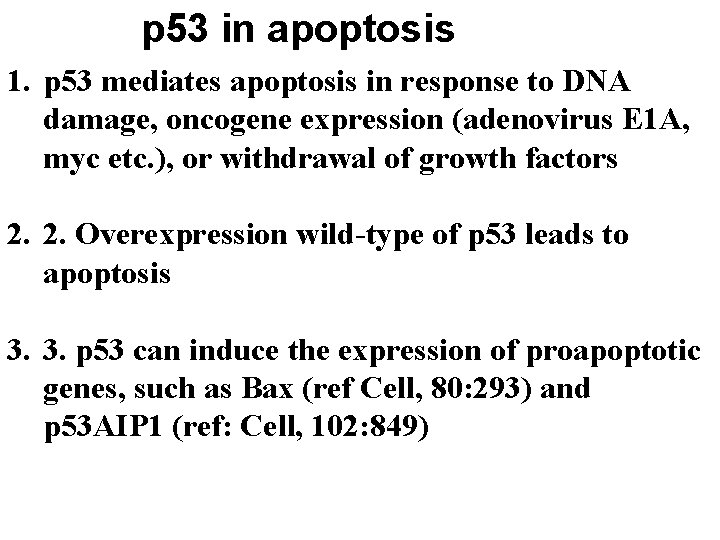
p 53 in apoptosis 1. p 53 mediates apoptosis in response to DNA damage, oncogene expression (adenovirus E 1 A, myc etc. ), or withdrawal of growth factors 2. 2. Overexpression wild-type of p 53 leads to apoptosis 3. 3. p 53 can induce the expression of proapoptotic genes, such as Bax (ref Cell, 80: 293) and p 53 AIP 1 (ref: Cell, 102: 849)

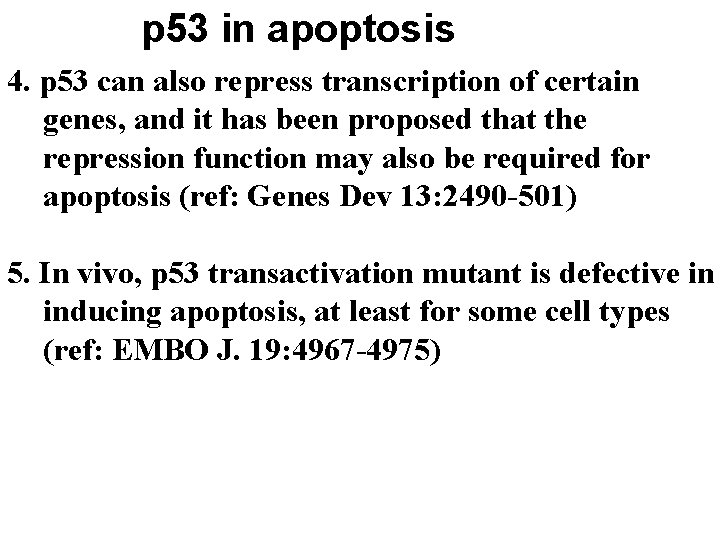
p 53 in apoptosis 4. p 53 can also repress transcription of certain genes, and it has been proposed that the repression function may also be required for apoptosis (ref: Genes Dev 13: 2490 -501) 5. In vivo, p 53 transactivation mutant is defective in inducing apoptosis, at least for some cell types (ref: EMBO J. 19: 4967 -4975)

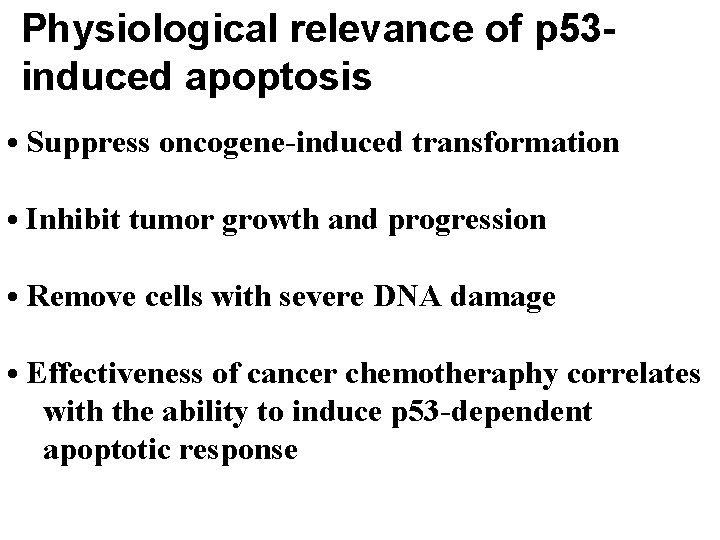
Physiological relevance of p 53 induced apoptosis • Suppress oncogene-induced transformation • Inhibit tumor growth and progression • Remove cells with severe DNA damage • Effectiveness of cancer chemotheraphy correlates with the ability to induce p 53 -dependent apoptotic response

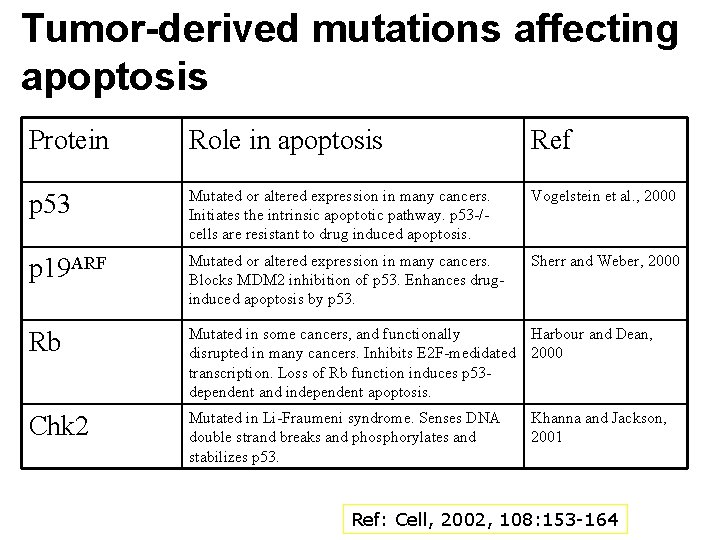
Tumor-derived mutations affecting apoptosis Protein Role in apoptosis Ref p 53 Mutated or altered expression in many cancers. Initiates the intrinsic apoptotic pathway. p 53 -/- cells are resistant to drug induced apoptosis. Vogelstein et al. , 2000 p 19 ARF Mutated or altered expression in many cancers. Blocks MDM 2 inhibition of p 53. Enhances druginduced apoptosis by p 53. Sherr and Weber, 2000 Rb Mutated in some cancers, and functionally Harbour and Dean, disrupted in many cancers. Inhibits E 2 F-medidated 2000 transcription. Loss of Rb function induces p 53 dependent and independent apoptosis. Chk 2 Mutated in Li-Fraumeni syndrome. Senses DNA double strand breaks and phosphorylates and stabilizes p 53. Khanna and Jackson, 2001 Ref: Cell, 2002, 108: 153 -164

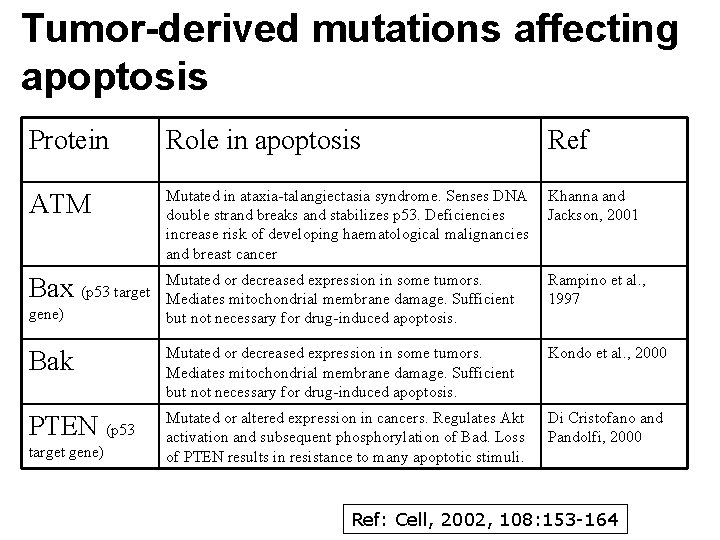
Tumor-derived mutations affecting apoptosis Protein Role in apoptosis ATM Mutated in ataxia-talangiectasia syndrome. Senses DNA Khanna and double strand breaks and stabilizes p 53. Deficiencies Jackson, 2001 increase risk of developing haematological malignancies and breast cancer Bax (p 53 target Mutated or decreased expression in some tumors. Mediates mitochondrial membrane damage. Sufficient but not necessary for drug-induced apoptosis. Rampino et al. , 1997 Bak Mutated or decreased expression in some tumors. Mediates mitochondrial membrane damage. Sufficient but not necessary for drug-induced apoptosis. Kondo et al. , 2000 PTEN (p 53 Mutated or altered expression in cancers. Regulates Akt activation and subsequent phosphorylation of Bad. Loss of PTEN results in resistance to many apoptotic stimuli. Di Cristofano and Pandolfi, 2000 gene) target gene) Ref: Cell, 2002, 108: 153 -164

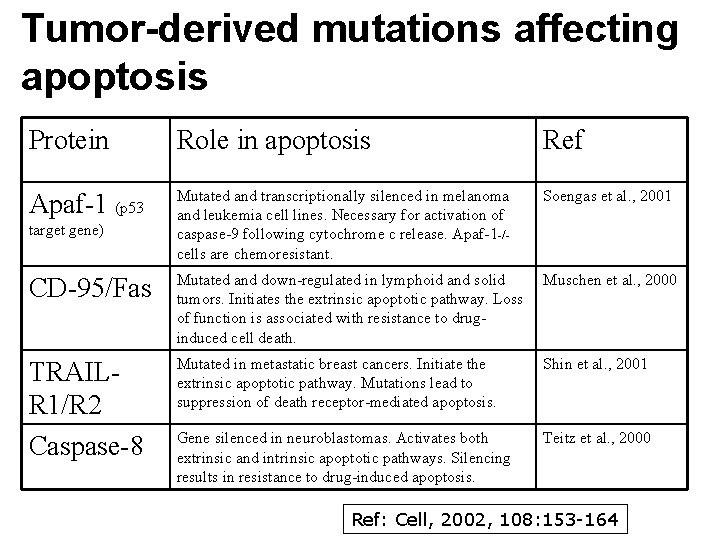
Tumor-derived mutations affecting apoptosis Protein Role in apoptosis Ref Apaf-1 (p 53 Mutated and transcriptionally silenced in melanoma and leukemia cell lines. Necessary for activation of caspase-9 following cytochrome c release. Apaf-1 -/- cells are chemoresistant. Soengas et al. , 2001 CD-95/Fas Mutated and down-regulated in lymphoid and solid tumors. Initiates the extrinsic apoptotic pathway. Loss of function is associated with resistance to druginduced cell death. Muschen et al. , 2000 TRAILR 1/R 2 Caspase-8 Mutated in metastatic breast cancers. Initiate the extrinsic apoptotic pathway. Mutations lead to suppression of death receptor-mediated apoptosis. Shin et al. , 2001 Gene silenced in neuroblastomas. Activates both extrinsic and intrinsic apoptotic pathways. Silencing results in resistance to drug-induced apoptosis. Teitz et al. , 2000 target gene) Ref: Cell, 2002, 108: 153 -164

Tumor-derived mutations affecting apoptosis Protein Role in apoptosis Ref Bcl 2 Frequently overexpressed in many tumors. Antagonises Reed, 1999 Bax and/or Bak and inhibits mitochondrial membrane disruption. Inhibits drug-induced apoptosis. MDM 2 Overexpressed in some tumors. Negative regulator of p 53. Inhibits drug-induced p 53 activation. Sherr and Weber, 2000 IAPs Frequently overexpressed in cancer. Down regulation of XIAP induces apoptosis in chemoresistant tumors. Deveraux and Reed, 1999 NFk. B Deregulated activity in many cancers. Transcriptionally Baldwin, 2001 activates expression of anti-apoptotic members of the Bcl-2 and IAP families. Can inhibit both the extrinsic and intrinsic death pathways and induce drugresistance. Ref: Cell, 2002, 108: 153 -164

Tumor-derived mutations affecting apoptosis Protein Role in apoptosis Ref Myc Deregulated expression in many cancers. Induces proliferation in the presence of survival factors, such as Bcl-2, and apoptosis in the absence of survival factors. Can sensitise cells to drug-induced apoptosis. Evan and Vousden, 2001 Akt Frequently amplified in solid tumors. Phosphorylates Bad. Hyperactivation induces resistance to a range of apoptotic stimuli including drugs. Datta et al. , 1999 PI 3 K Overexpressed or deregulated in some cancers. Responsible for activation of Akt and downstream phosphorylation of Bad. Inhibition of PI 3 K enhances chemotherapeutic drug-induced apoptosis. Roymans and Slegers, 2001 Ras Mutated or deregulated in many cancers. Activates PI 3 K and downstream pathways. Induces proliferation and inhibits c-myc and drug-induced apoptosis. el-Deiry, 1997 Ref: Cell, 2002, 108: 153 -164

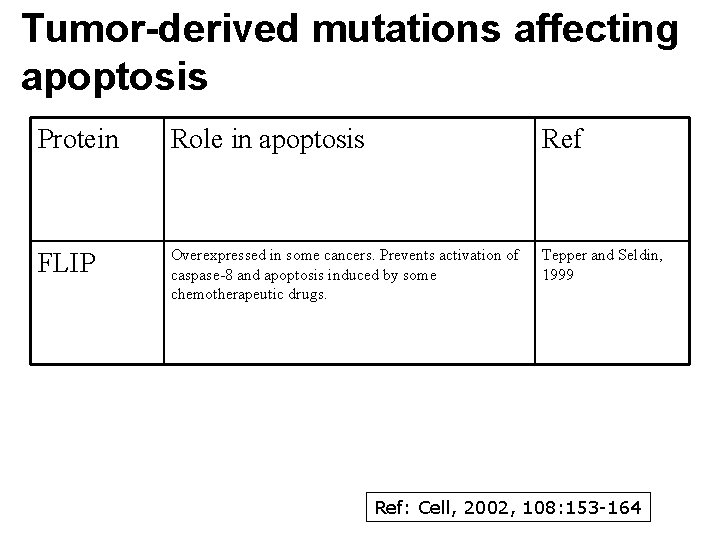
Tumor-derived mutations affecting apoptosis Protein Role in apoptosis Ref FLIP Overexpressed in some cancers. Prevents activation of caspase-8 and apoptosis induced by some chemotherapeutic drugs. Tepper and Seldin, 1999 Ref: Cell, 2002, 108: 153 -164

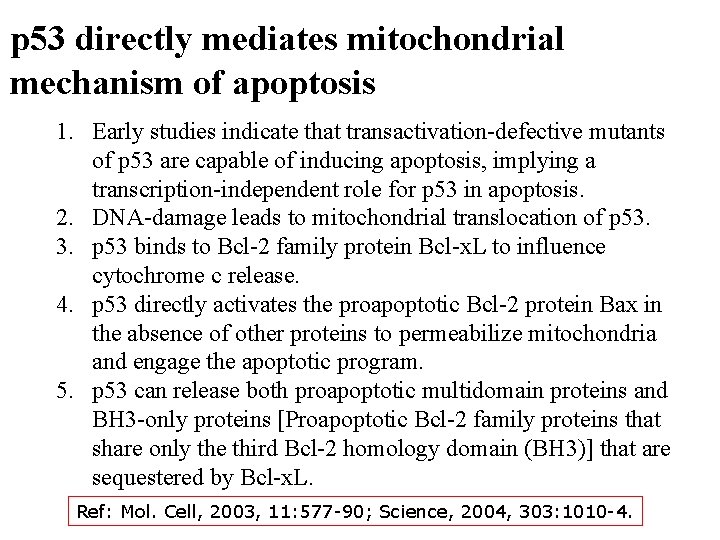
p 53 directly mediates mitochondrial mechanism of apoptosis 1. Early studies indicate that transactivation-defective mutants of p 53 are capable of inducing apoptosis, implying a transcription-independent role for p 53 in apoptosis. 2. DNA-damage leads to mitochondrial translocation of p 53. 3. p 53 binds to Bcl-2 family protein Bcl-x. L to influence cytochrome c release. 4. p 53 directly activates the proapoptotic Bcl-2 protein Bax in the absence of other proteins to permeabilize mitochondria and engage the apoptotic program. 5. p 53 can release both proapoptotic multidomain proteins and BH 3 -only proteins [Proapoptotic Bcl-2 family proteins that share only the third Bcl-2 homology domain (BH 3)] that are sequestered by Bcl-x. L. Ref: Mol. Cell, 2003, 11: 577 -90; Science, 2004, 303: 1010 -4.

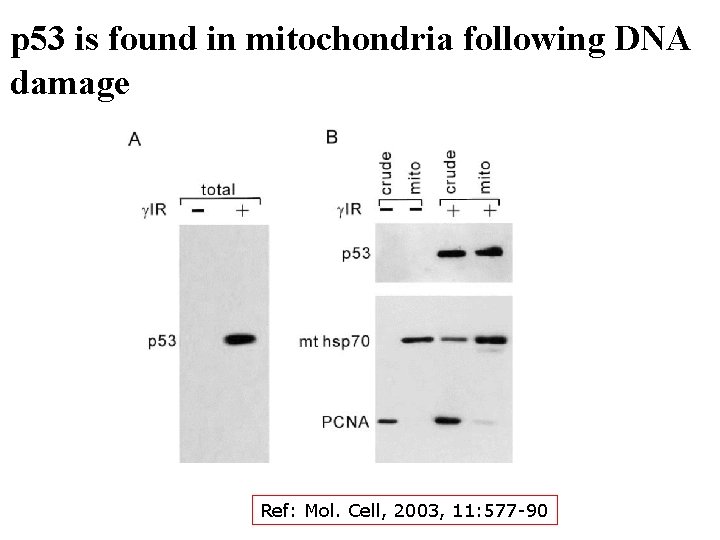
p 53 is found in mitochondria following DNA damage Ref: Mol. Cell, 2003, 11: 577 -90

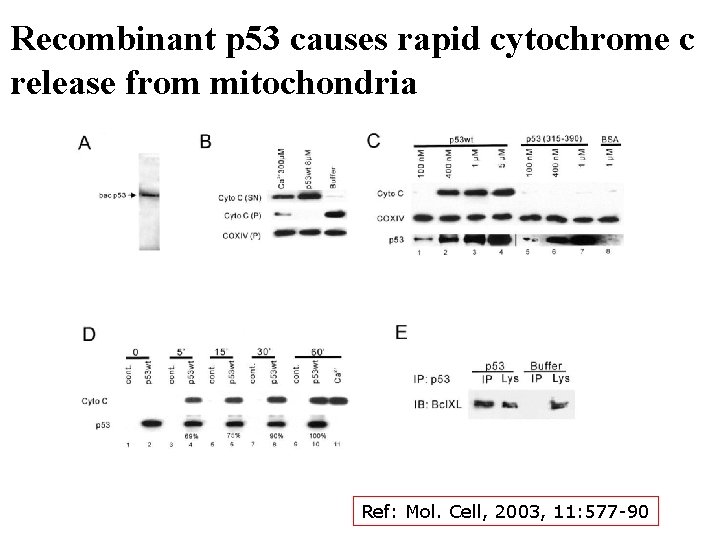
Recombinant p 53 causes rapid cytochrome c release from mitochondria Ref: Mol. Cell, 2003, 11: 577 -90

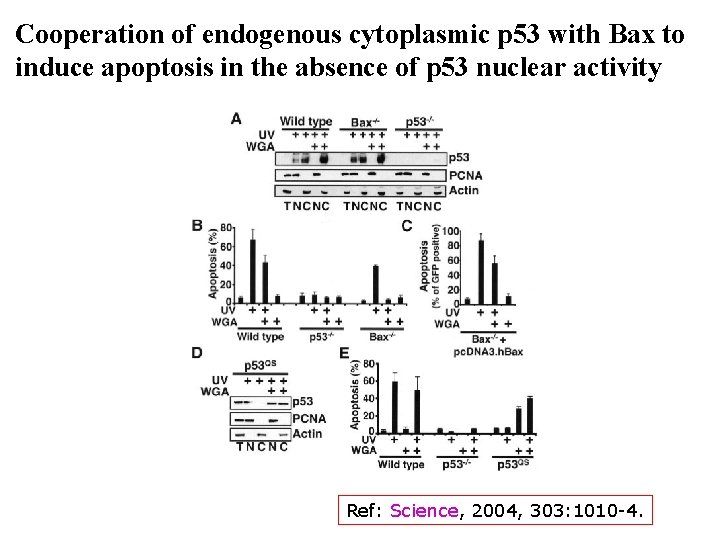
Cooperation of endogenous cytoplasmic p 53 with Bax to induce apoptosis in the absence of p 53 nuclear activity Ref: Science, 2004, 303: 1010 -4.

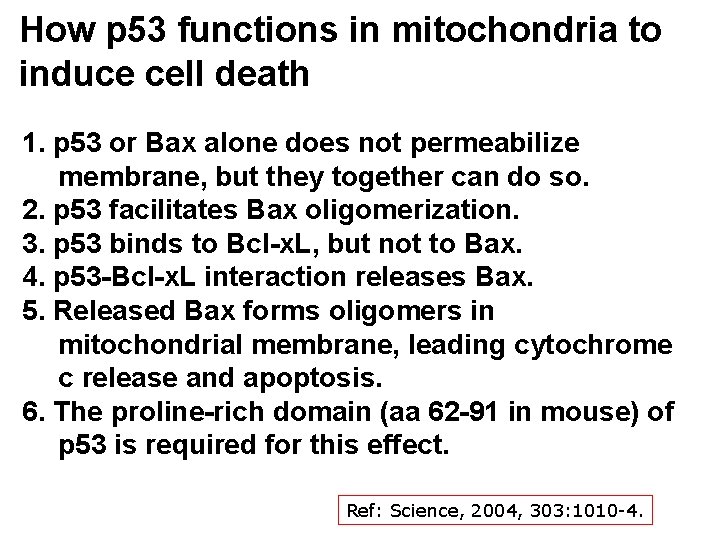
How p 53 functions in mitochondria to induce cell death 1. p 53 or Bax alone does not permeabilize membrane, but they together can do so. 2. p 53 facilitates Bax oligomerization. 3. p 53 binds to Bcl-x. L, but not to Bax. 4. p 53 -Bcl-x. L interaction releases Bax. 5. Released Bax forms oligomers in mitochondrial membrane, leading cytochrome c release and apoptosis. 6. The proline-rich domain (aa 62 -91 in mouse) of p 53 is required for this effect. Ref: Science, 2004, 303: 1010 -4.

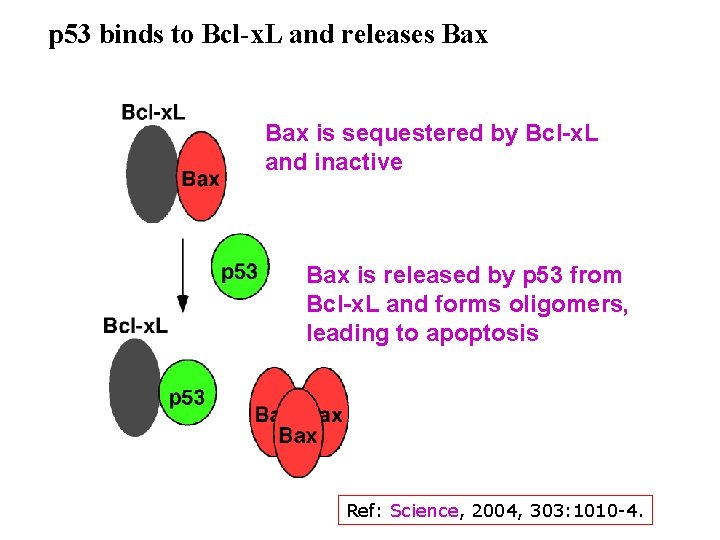
p 53 binds to Bcl-x. L and releases Bax is sequestered by Bcl-x. L and inactive Bax is released by p 53 from Bcl-x. L and forms oligomers, leading to apoptosis Ref: Science, 2004, 303: 1010 -4.

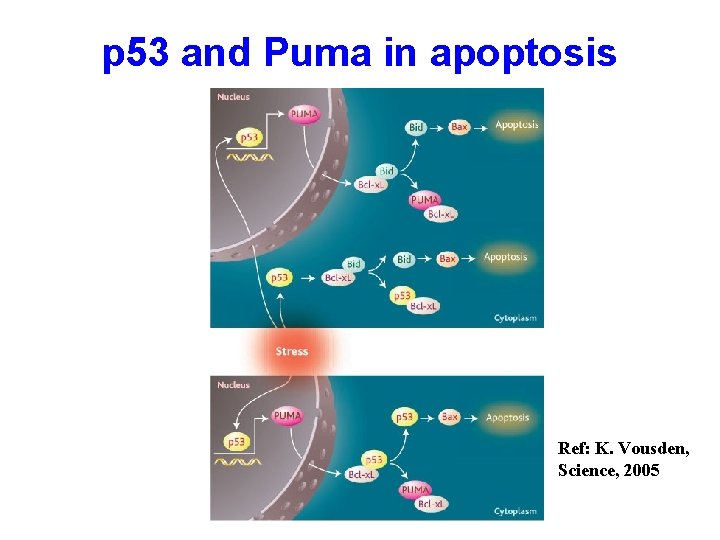
p 53 and Puma in apoptosis Ref: K. Vousden, Science, 2005

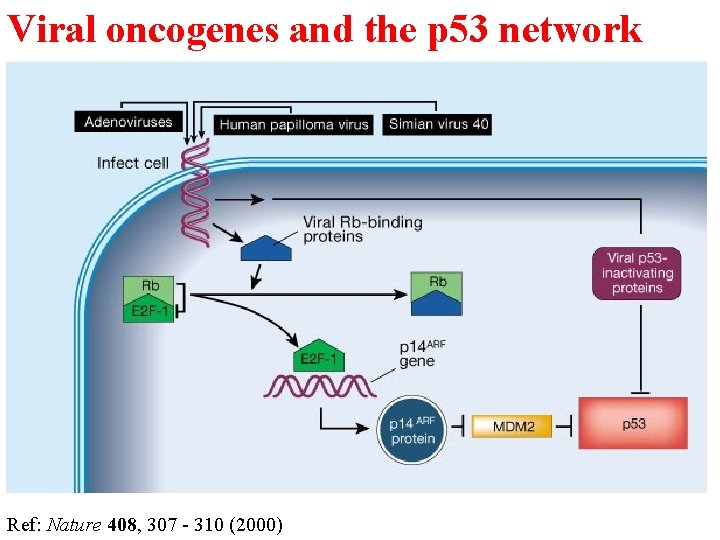
Viral oncogenes and the p 53 network Ref: Nature 408, 307 - 310 (2000)

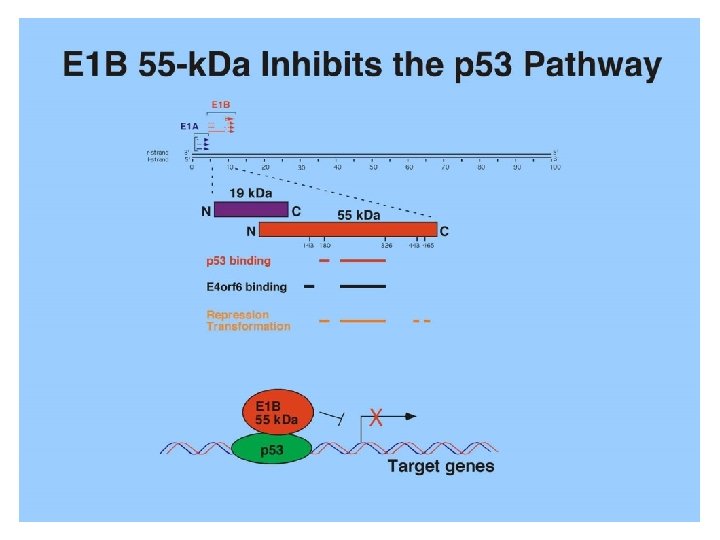
Viral oncogenes inhibit p 53 functions 1. p 53 was first identified as binding protein of SV 40 large T antigen in 1979, which interacts with p 53 core domain 2. Adenovirus oncoprotein E 1 B 55 -k. Da binds to p 53 and inhibits p 53 transactivation activity 3. HPV E 6 binds to p 53 and targets it for ubiquitinmediated degradation.



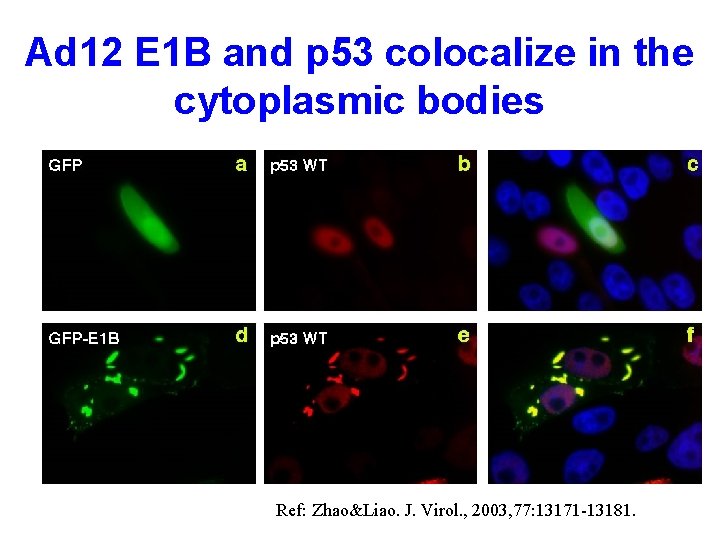
Ad 12 E 1 B and p 53 colocalize in the cytoplasmic bodies Ref: Zhao&Liao. J. Virol. , 2003, 77: 13171 -13181.

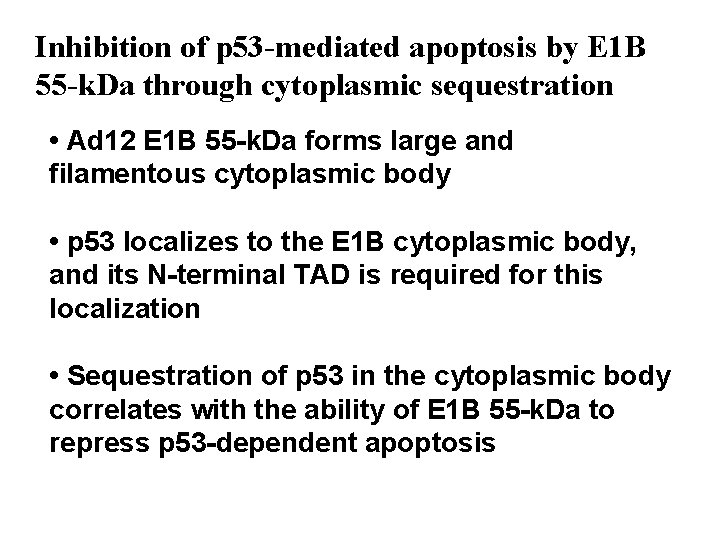
Inhibition of p 53 -mediated apoptosis by E 1 B 55 -k. Da through cytoplasmic sequestration • Ad 12 E 1 B 55 -k. Da forms large and filamentous cytoplasmic body • p 53 localizes to the E 1 B cytoplasmic body, and its N-terminal TAD is required for this localization • Sequestration of p 53 in the cytoplasmic body correlates with the ability of E 1 B 55 -k. Da to repress p 53 -dependent apoptosis

Using p 53 to kill cancer cells The p 53 protein is a tumor suppressor — it keeps cell numbers down by stopping cells from multiplying or by promoting cell death. Loss of p 53 occurs in most human cancers, so it would be useful to be able to restore its function. Several innovative strategies have been suggested: • Introduce normal p 53 genes into a cancer cell with mutant p 53. • Introduce a small compound that converts mutant p 53 proteins from an abnormal to a normal shape. • Add a protein that attaches itself to mutant p 53 and kills cells. • Stimulate the host's immune response to mutant p 53 peptides.

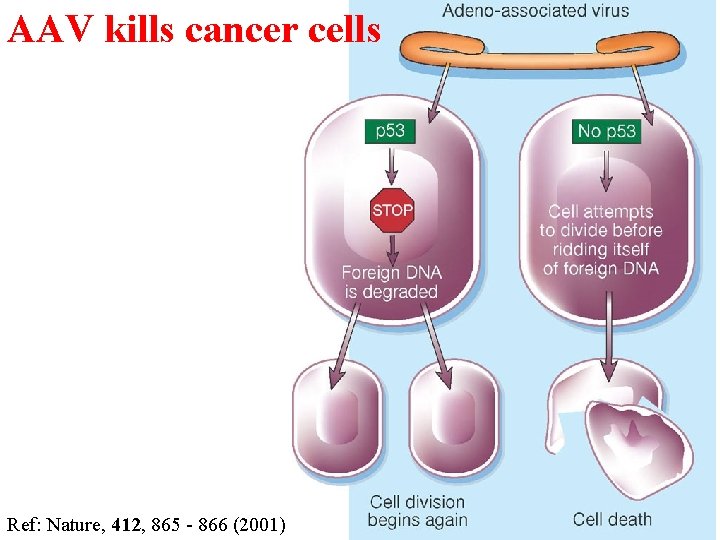
Using p 53 to kill cancer cells • Introduce drugs that disrupt the interaction between the MDM 2 or E 6 proteins and p 53. (MDM 2 and E 6 negatively regulate p 53; they are present at abnormally high levels in some cancer cells, so 'quench' any normal p 53. ) • Introduce the adeno-associated virus, which mimics damaged DNA. Cells with mutant p 53 cannot activate the usual p 53 -dependent 'checkpoint' that is induced by DNA damage, and eventually die. • Infect cells with viruses that can replicate only in cells without normal p 53; the viruses kill these cells.

AAV kills cancer cells Ref: Nature, 412, 865 - 866 (2001)

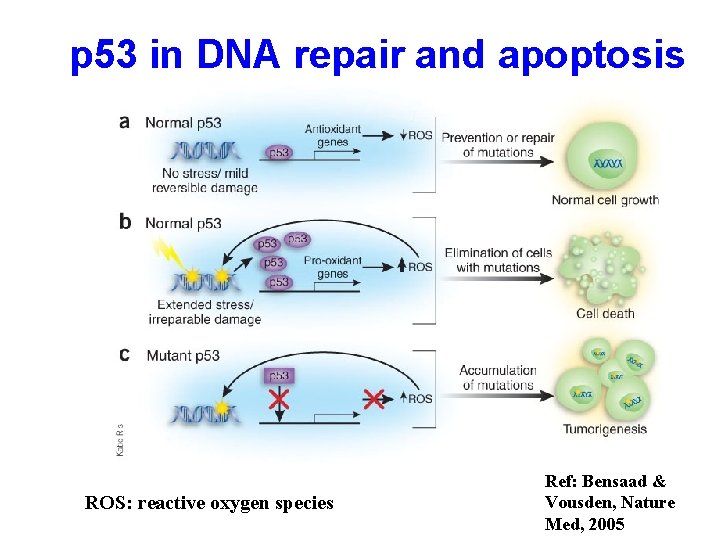
p 53 in DNA repair and apoptosis ROS: reactive oxygen species Ref: Bensaad & Vousden, Nature Med, 2005

Summary 1. p 53 is a tumor-suppressor protein that induces apoptotic cell death in response to oncogenic stress. Malignant progression is dependent on loss of p 53 function, either through mutation in the TP 53 gene (which encodes p 53) itself or by defects in the signaling pathways that are upstream or downstream of p 53. 2. Mutations in TP 53 occur in about half of all human cancers, almost always resulting in the expression of a mutant p 53 protein that has acquired transforming activity. 3. p 53 -induced apoptosis depends on the ability of p 53 to activate gene expression. 4. p 53 can also directly trigger the apoptotic response, by interacting with Bcl-2 family protein. 5. The apoptotic and cell-cycle arrest activities of p 53 can be separated, and apoptotic cofactors that play a specific role in allowing p 53 -induced death are being identified. Ref: Nature Reviews Cancer, 2, 594 - 604 (2002)

Summary 6. Phosphorylation of p 53 regulates its ability to activate the expression of apoptotic target genes, and other posttranslational modifications such as acetylation might also have a role. 7. In tumors that retain wild-type p 53, the apoptotic response might be hindered by defects in the apoptotic cofactors. These, therefore, represent additional targets for the design of therapeutics that are aimed at reactivating p 53 -mediated apoptosis in cancer cells. Ref: Nature Reviews Cancer, 2, 594 - 604 (2002)