Geometric Algorithms for Conformational Analysis of Long Protein



- Slides: 23

Geometric Algorithms for Conformational Analysis of Long Protein Loops J. Cortess, T. Simeon, M. Remaud. Simeon, V. Tran

Motivation Filter unfeasible loop conformations to aid searching conformational space for various application: – Protein loop modeling – Molecular simulations: conformational changes under environmental conditions.

Structural Constraints • Loop-closure • Steric clash – internal segment clashes (selfclashes), external clashes, Vd. W radii.

Loop Closure Approaches • Analytical – IK techniques • Optimization – e. g. CCD • Databased methods

Clash Filtering Approaches • Energetic – accepting/rejecting a conformation according to some energetic (repulsive Vd. W energy) cutoff. • Geometric – “clash grids”. • Robotics – motion planning.

Robotics – collision avoidance Exploration of the conformations space, searching for feasible conformations. Existing techniques capture the topology of the feasible space within a data-structure (graph or a tree) by performing random exploration.

Outline • Part 1: presents conformational sampling technique satisfying loop-closure and clash avoidance constraints. • Part 2: presents a data structure capturing the connectivity of the geometrically feasible conformations sub-space.

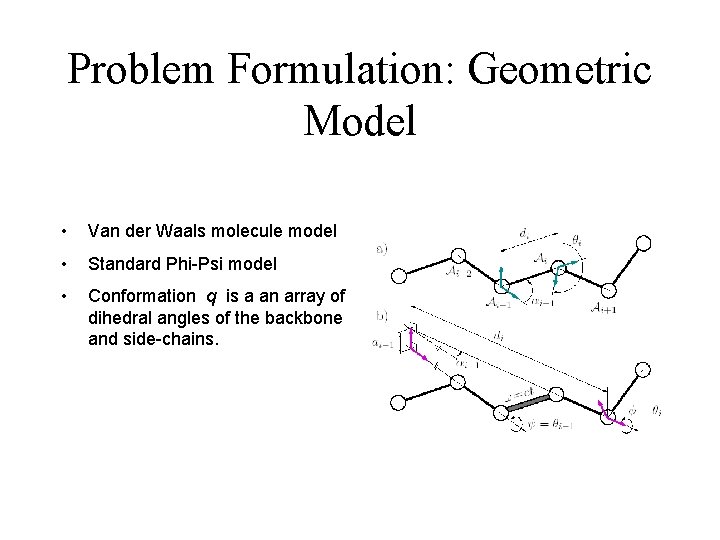
Problem Formulation: Geometric Model • Van der Waals molecule model • Standard Phi-Psi model • Conformation q is a an array of dihedral angles of the backbone and side-chains.

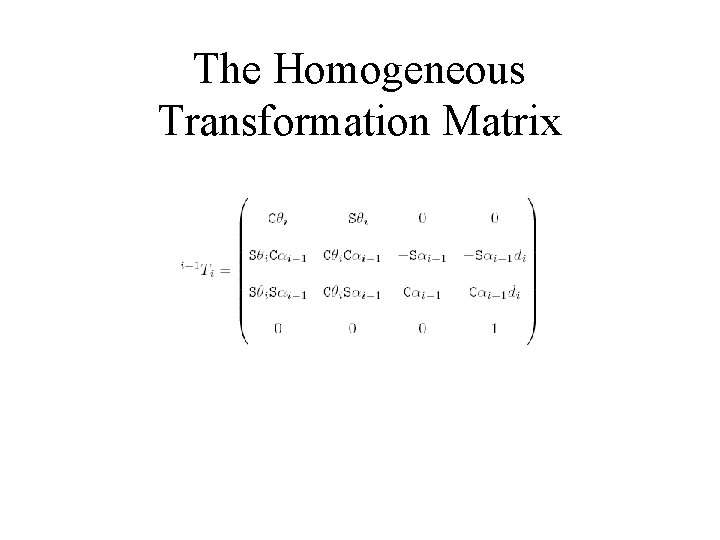
The Homogeneous Transformation Matrix

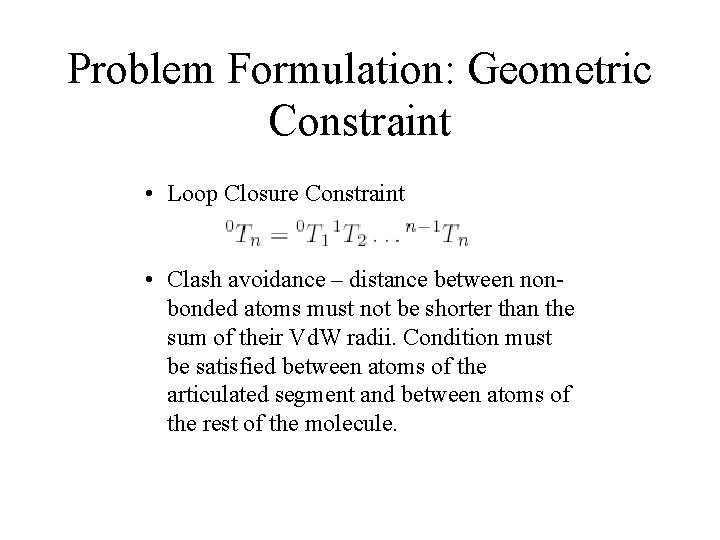
Problem Formulation: Geometric Constraint • Loop Closure Constraint • Clash avoidance – distance between nonbonded atoms must not be shorter than the sum of their Vd. W radii. Condition must be satisfied between atoms of the articulated segment and between atoms of the rest of the molecule.

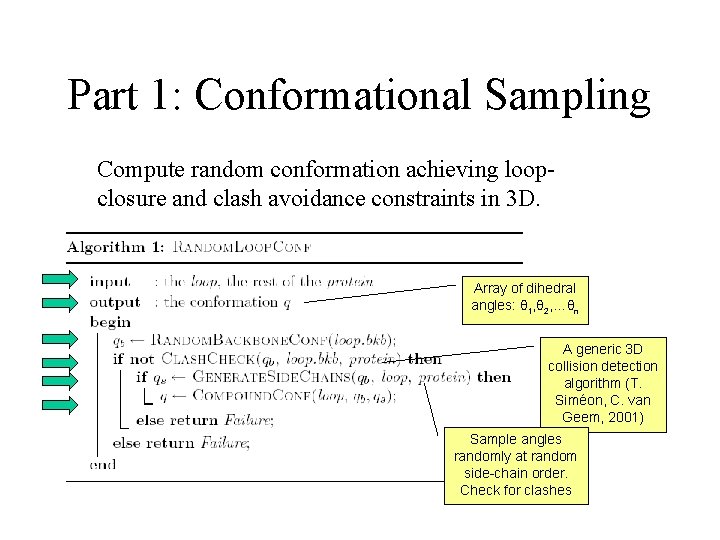
Part 1: Conformational Sampling Compute random conformation achieving loopclosure and clash avoidance constraints in 3 D. Array of dihedral angles: θ 1, θ 2, …θn A generic 3 D collision detection algorithm (T. Siméon, C. van Geem, 2001) Sample angles randomly at random side-chain order. Check for clashes

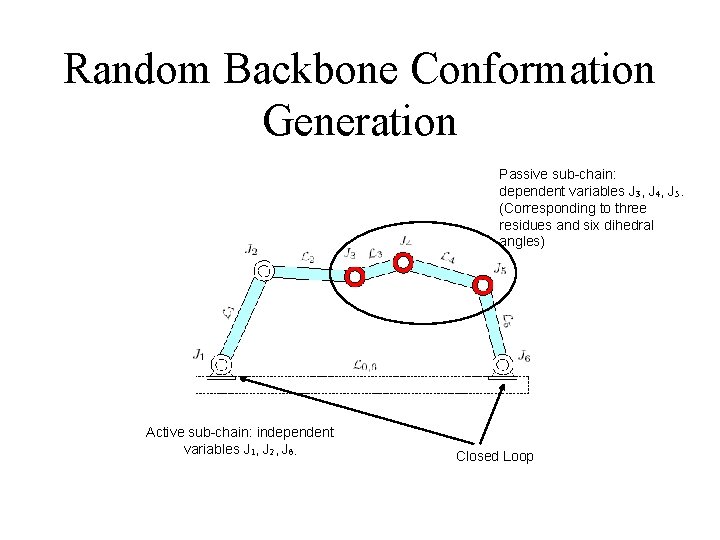
Random Backbone Conformation Generation Passive sub-chain: dependent variables J 3, J 4, J 5. (Corresponding to three residues and six dihedral angles) Active sub-chain: independent variables J 1, J 2, J 6. Closed Loop

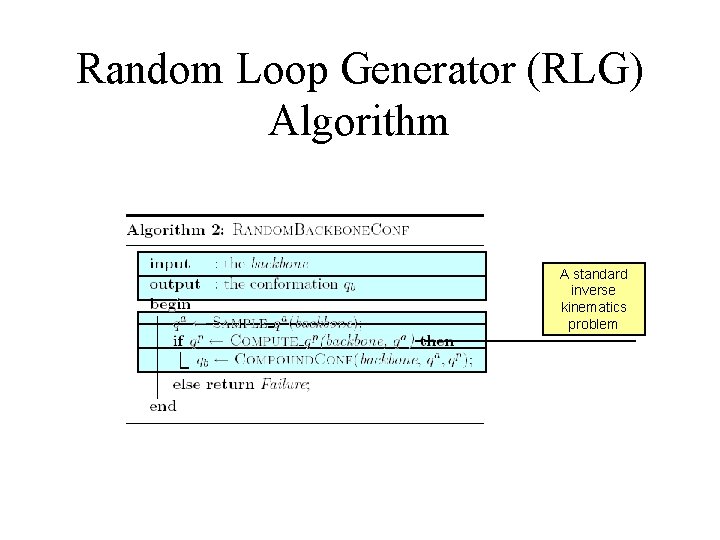
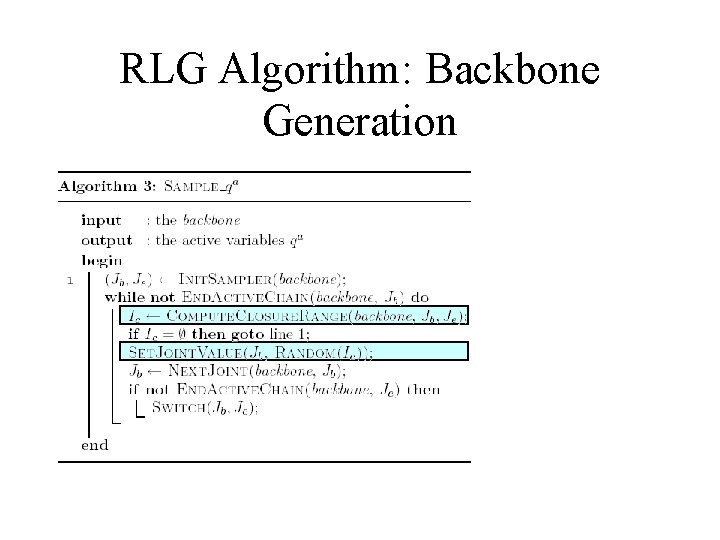
Random Loop Generator (RLG) Algorithm A standard inverse kinematics problem

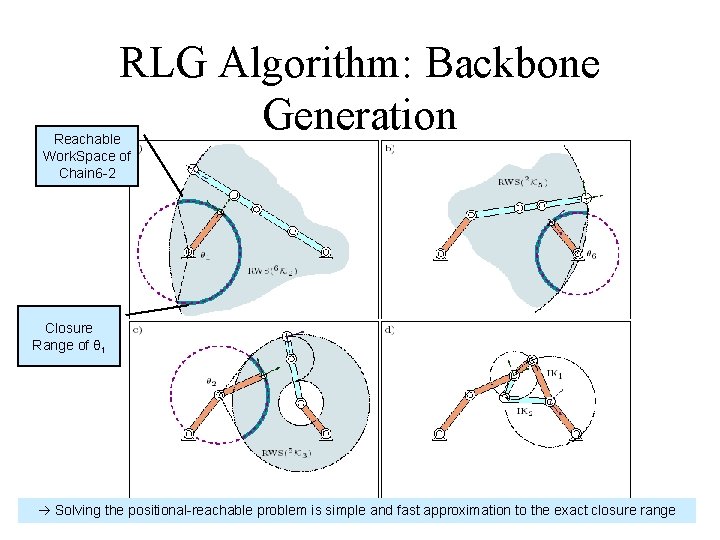
RLG Algorithm: Backbone Generation Reachable Work. Space of Chain 6 -2 Closure Range of θ 1 Solving the positional-reachable problem is simple and fast approximation to the exact closure range

RLG Algorithm: Backbone Generation

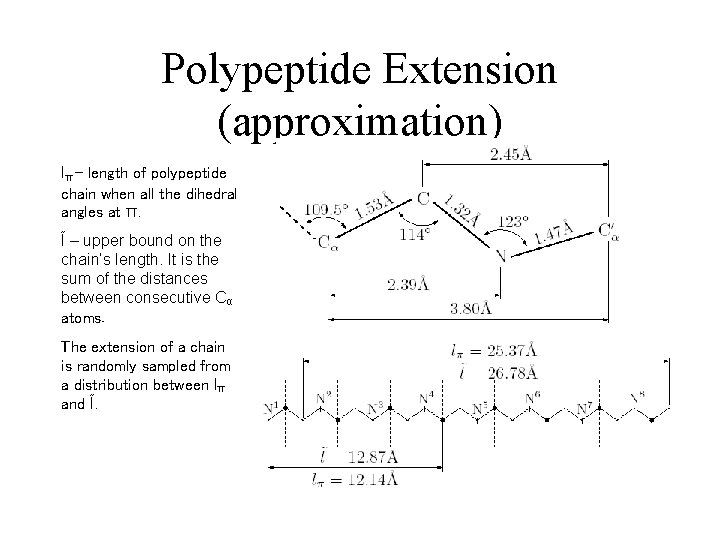
Polypeptide Extension (approximation) lπ – length of polypeptide chain when all the dihedral angles at π. Ĩ – upper bound on the chain’s length. It is the sum of the distances between consecutive Cα atoms. The extension of a chain is randomly sampled from a distribution between lπ and Ĩ.

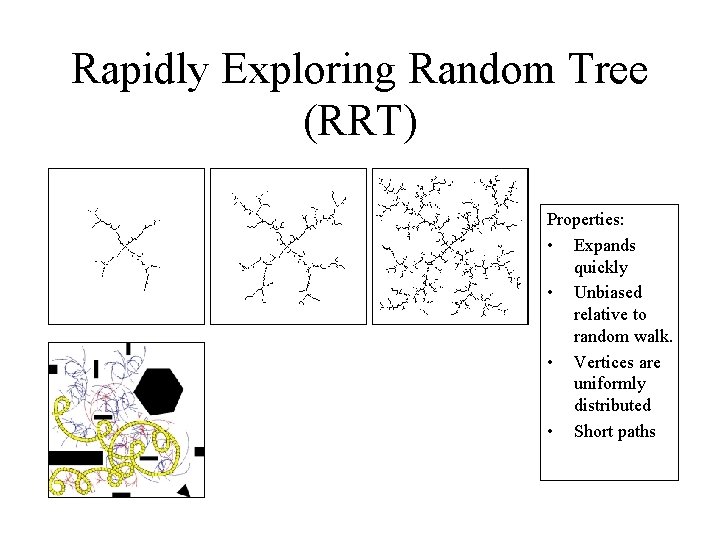
Part 2: Conformational Space Exploration Apply Sampling-based Motion Planning Techniques to the Protein Loop Problem. In particular, the Probabilistic Road. Map (PRM) approach. Rapidly-exploring Random Tree (RRT) is a data structure and a sampling scheme to quickly search high-dimensional constrained spaces.

Rapidly Exploring Random Tree (RRT) Properties: • Expands quickly • Unbiased relative to random walk. • Vertices are uniformly distributed • Short paths

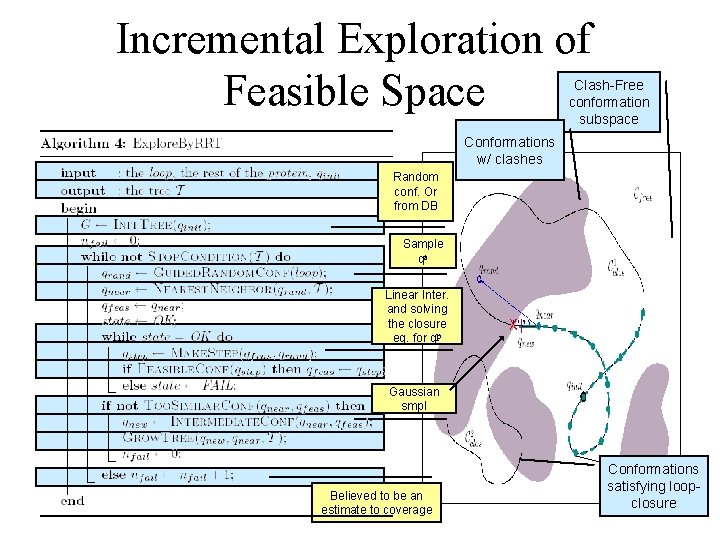
Incremental Exploration of Feasible Space Clash-Free conformation subspace Conformations w/ clashes Random conf. Or from DB Sample qa Linear Inter. and solving the closure eq. for qp Gaussian smpl Believed to be an estimate to coverage Conformations satisfying loopclosure

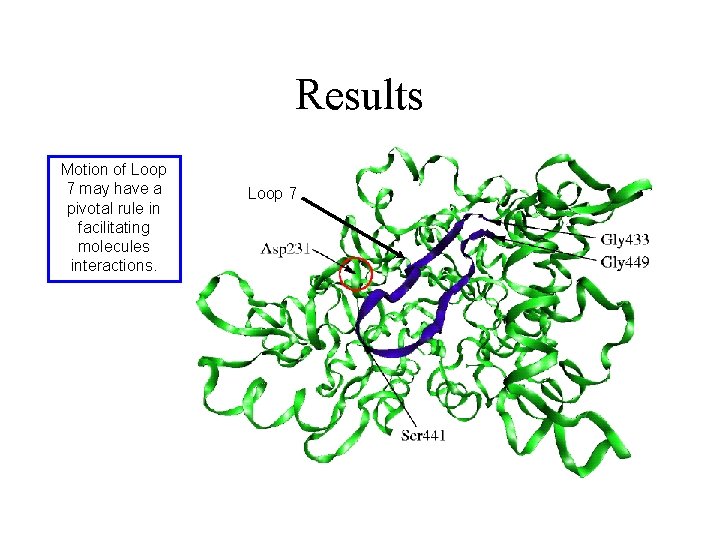
Results Motion of Loop 7 may have a pivotal rule in facilitating molecules interactions. Loop 7

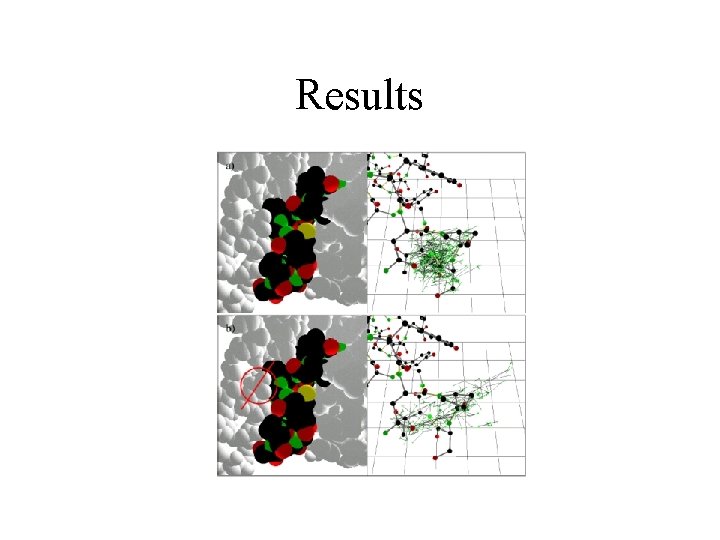
Results

CCD vs. RLG • Similar performance in terms of finding conformations close to the wild-type. • RLG computes exact solutions while CCD outputs approximated solutions. • CCD may favor large changes in the first residues. RLG produces a more uniformly distributed samples.

Future Directions • Check clashes at each stage. • Tailor a collision detection algorithm for the molecular application (Collision detection is by far the most computation expensive task) • Incorporate energetic analysis (constraints) into the incremental search technique.