Genomics of Gene Regulation Genomic and Proteomic Approaches













- Slides: 53

Genomics of Gene Regulation Genomic and Proteomic Approaches to Heart, Lung, Blood and Sleep Disorders Jackson Laboratories Ross Hardison September 9, 2008

Heritable variation in gene regulation “Simple” Mendelian traits, e. g. thalassemias Variation in expression is common in normal individuals Variation in expression may be a major contributor to complex traits (including heart, lung, blood and sleep disorders)

Deletions of noncoding DNA can affect gene expression Forget and Hardison, Chapter in Disorders of Hemoglobin, 2 nd edition

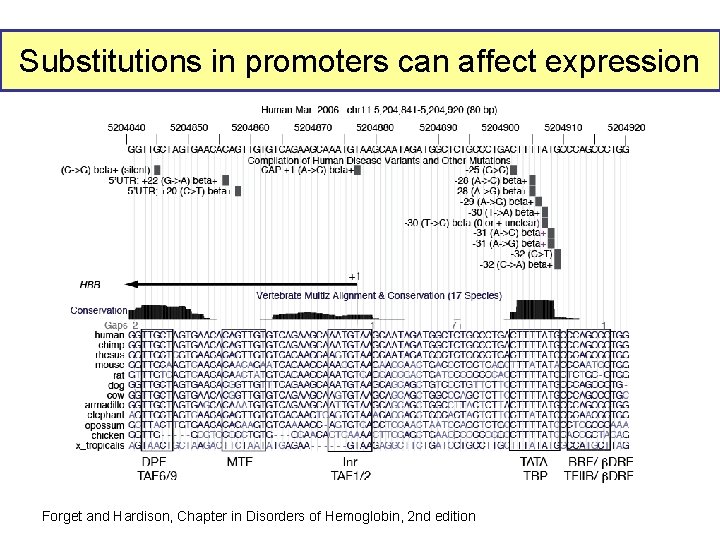
Substitutions in promoters can affect expression Forget and Hardison, Chapter in Disorders of Hemoglobin, 2 nd edition

Variation of gene expression among individuals • Levels of expression of many genes varies in humans (and other species) • Variation in expression is heritable • Determinants of variability map to discrete genomic intervals • Often multiple determinants • Points to an abundance of cis-regulatory variation in the human genome • "We predict that variants in regulatory regions make a greater contribution to complex disease than do variants that affect protein sequence" Manolis Dermitzakis, Science. Daily – Microarray expression analyses of 3554 genes in 14 families • Morley M … Cheung VG (2004) Nature 430: 743 -747 – Expression analysis of EBV-transformed lymphoblastoid cells from all 270 individuals genotypes in Hap. Map • Stranger BE … Dermitzakis E (2007) Nature Genetics 39: 1217 -1224

Risk loci in noncoding regions (2007) Science 316: 1336 -1341

DNA sequences involved in regulation of gene transcription Protein-DNA interactions Chromatin effects

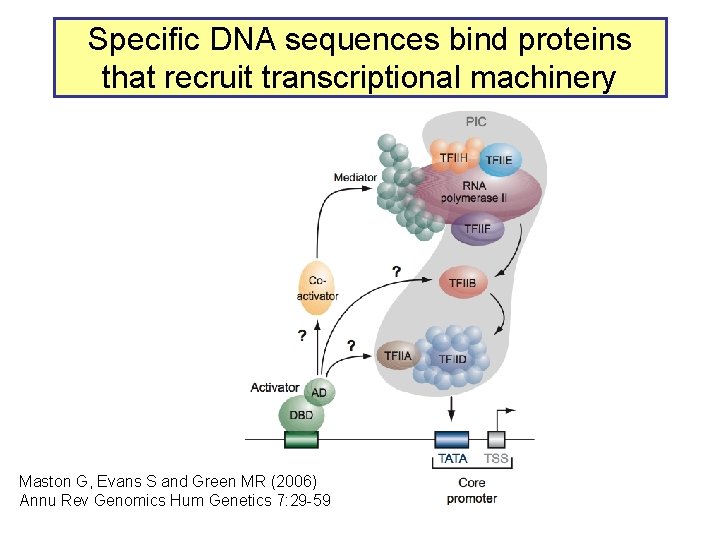
Specific DNA sequences bind proteins that recruit transcriptional machinery Maston G, Evans S and Green MR (2006) Annu Rev Genomics Hum Genetics 7: 29 -59

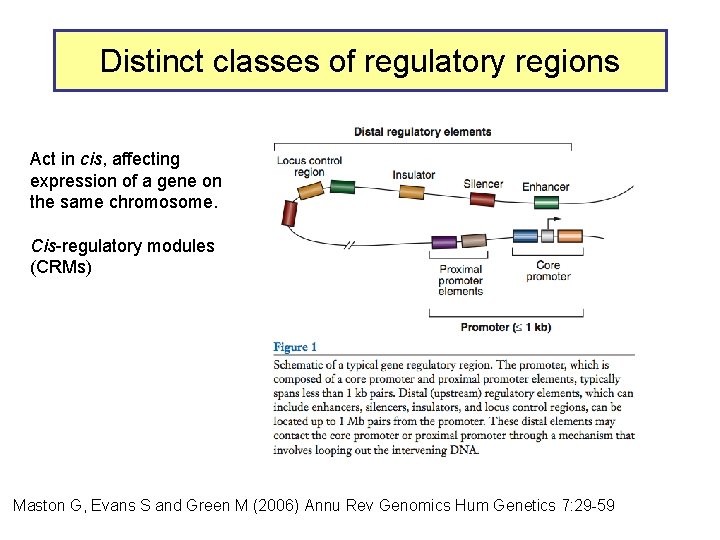
Distinct classes of regulatory regions Act in cis, affecting expression of a gene on the same chromosome. Cis-regulatory modules (CRMs) Maston G, Evans S and Green M (2006) Annu Rev Genomics Hum Genetics 7: 29 -59

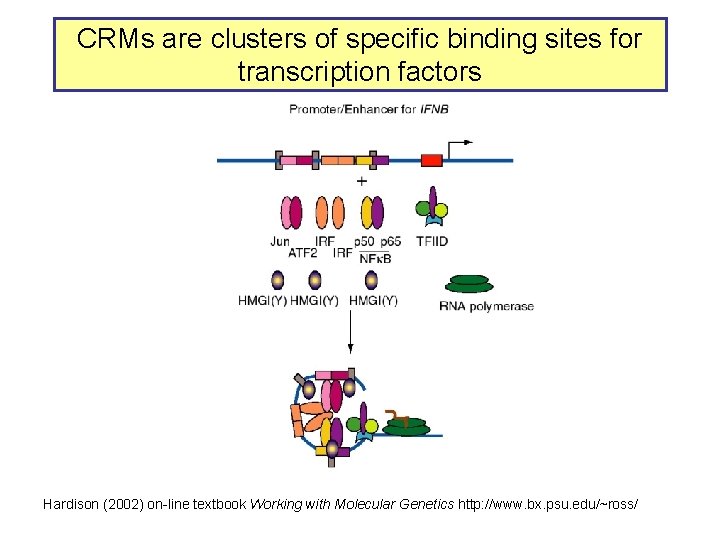
CRMs are clusters of specific binding sites for transcription factors Hardison (2002) on-line textbook Working with Molecular Genetics http: //www. bx. psu. edu/~ross/

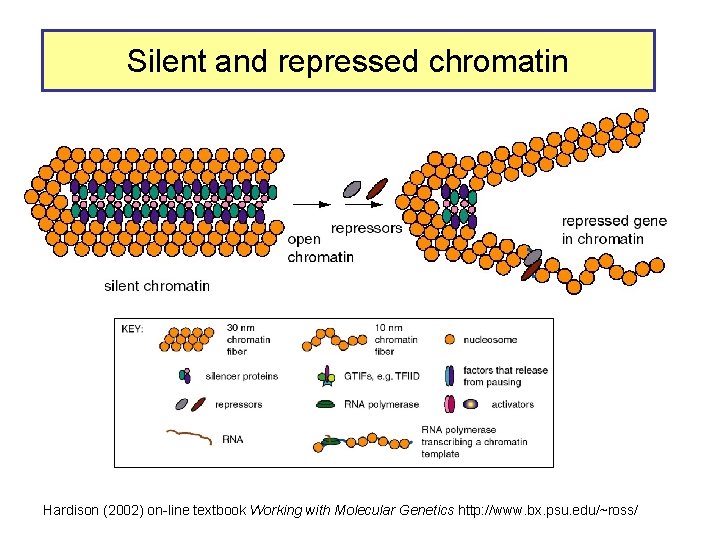
Silent and repressed chromatin Hardison (2002) on-line textbook Working with Molecular Genetics http: //www. bx. psu. edu/~ross/

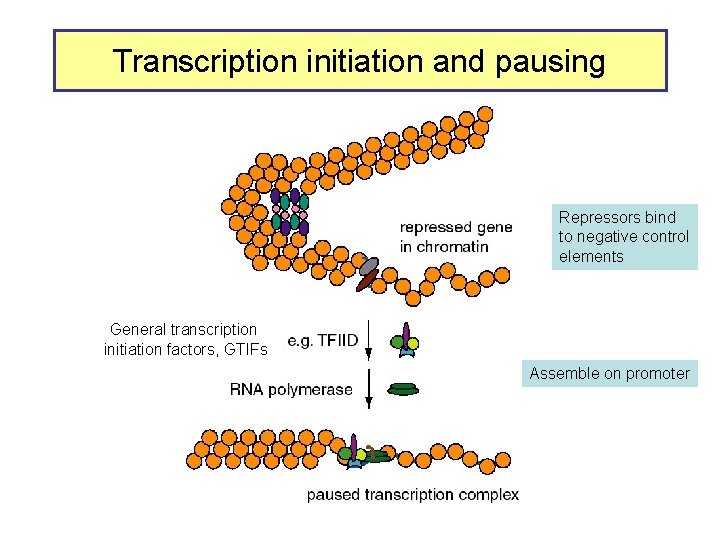
Transcription initiation and pausing Repressors bind to negative control elements General transcription initiation factors, GTIFs Assemble on promoter

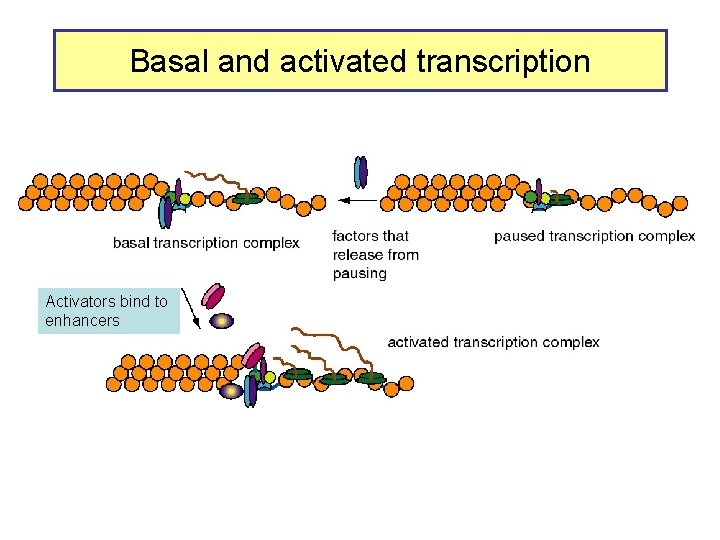
Basal and activated transcription Activators bind to enhancers

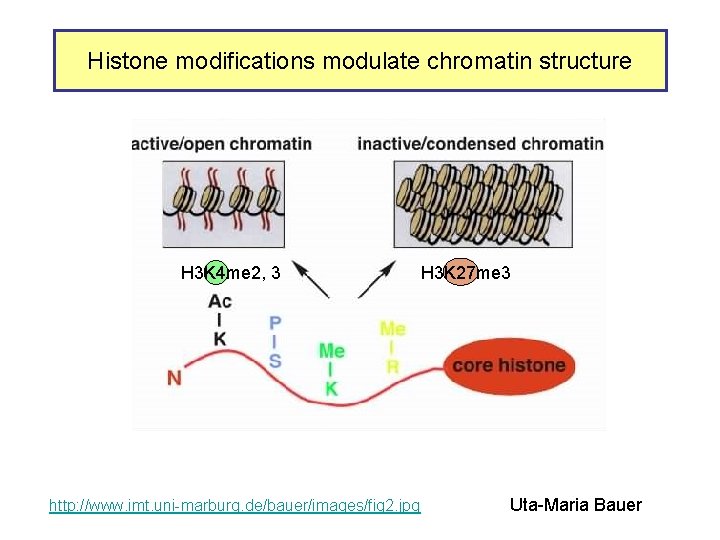
Histone modifications modulate chromatin structure H 3 K 4 me 2, 3 http: //www. imt. uni-marburg. de/bauer/images/fig 2. jpg H 3 K 27 me 3 Uta-Maria Bauer

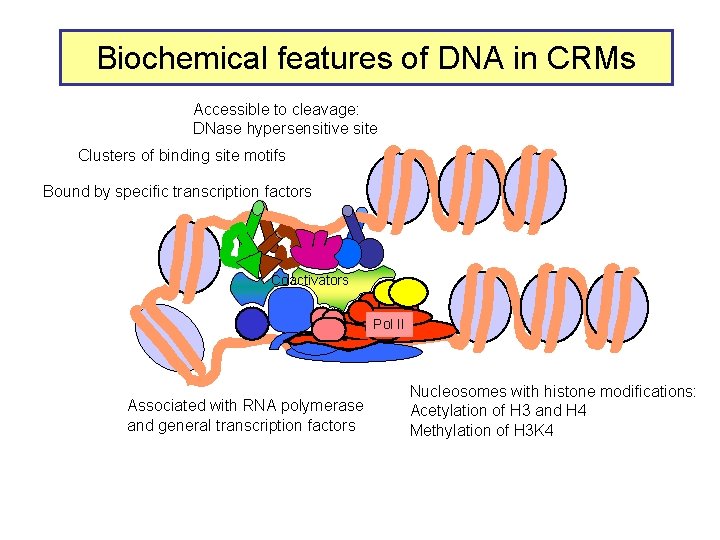
Biochemical features of DNA in CRMs Accessible to cleavage: DNase hypersensitive site Clusters of binding site motifs Bound by specific transcription factors Coactivators Pol IIa II Associated with RNA polymerase and general transcription factors Nucleosomes with histone modifications: Acetylation of H 3 and H 4 Methylation of H 3 K 4

Examples of genome-wide data on CRM features • RNA polymerase II, preinitiation complex – IMR 90 cells: Kim TH …Ren B (2005) Nature 436: 876 -880 • Start sites for transcription – Carninci et al. (2006) Nature Genetics 38: 626 -635 • Histone modifications – T cells: Roh. . . Zhao K (2006) PNAS 103: 15782 -15878 • Insulator protein CTCF – Primary fibroblasts: Kim TH … Ren B (2007) Cell 128: 1231 -1245 • DNase hypersensitive sites – CD 4+ T cells: Boyle… Crawford G (2008) Cell 132: 311 -322 • Many datastreams: ENCODE project – Birney et al. (2007) Nature 477: 799 -816

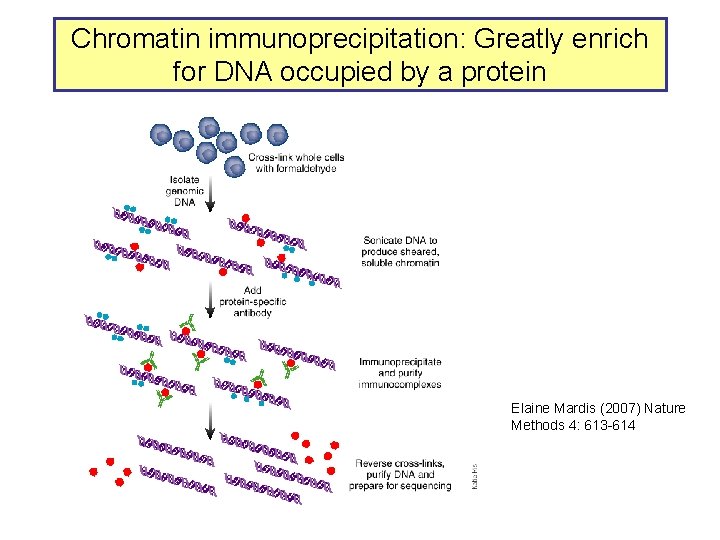
Chromatin immunoprecipitation: Greatly enrich for DNA occupied by a protein Elaine Mardis (2007) Nature Methods 4: 613 -614

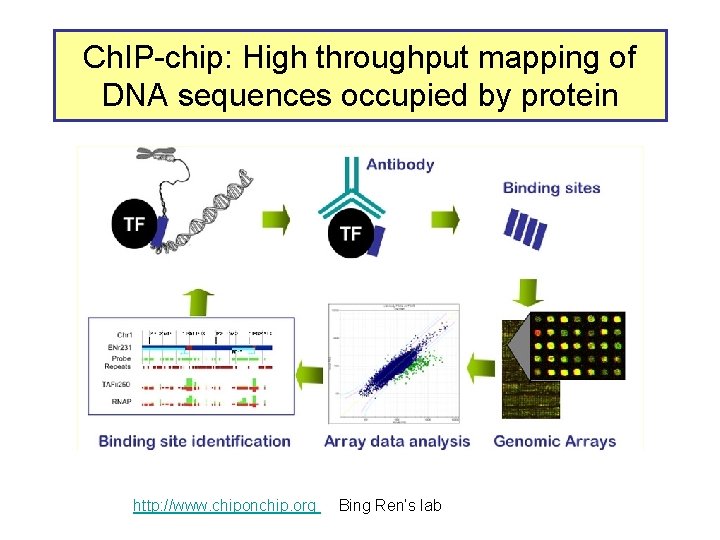
Ch. IP-chip: High throughput mapping of DNA sequences occupied by protein http: //www. chiponchip. org Bing Ren’s lab

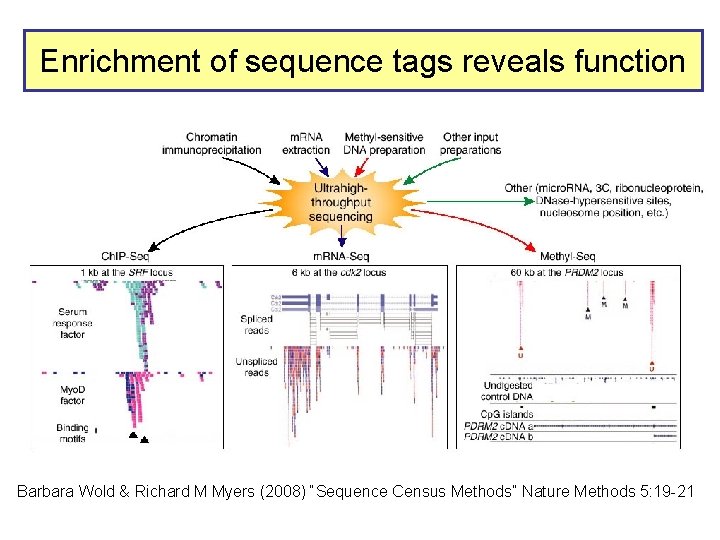
Enrichment of sequence tags reveals function Barbara Wold & Richard M Myers (2008) “Sequence Census Methods” Nature Methods 5: 19 -21

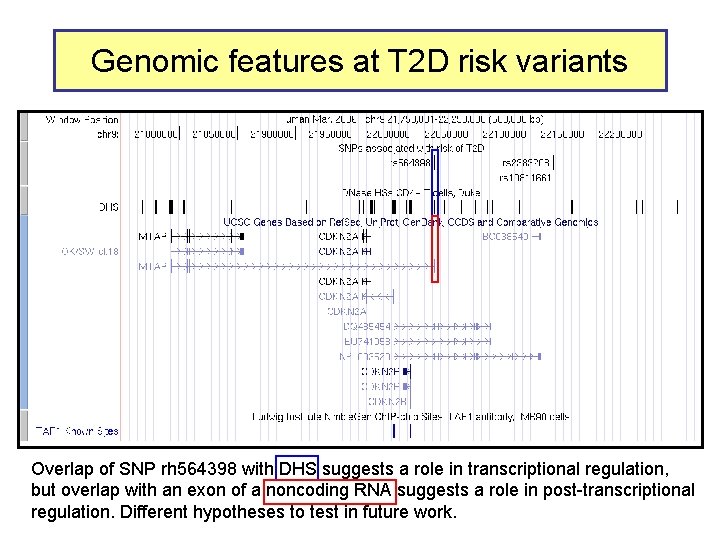
Genomic features at T 2 D risk variants Overlap of SNP rh 564398 with DHS suggests a role in transcriptional regulation, but overlap with an exon of a noncoding RNA suggests a role in post-transcriptional regulation. Different hypotheses to test in future work.

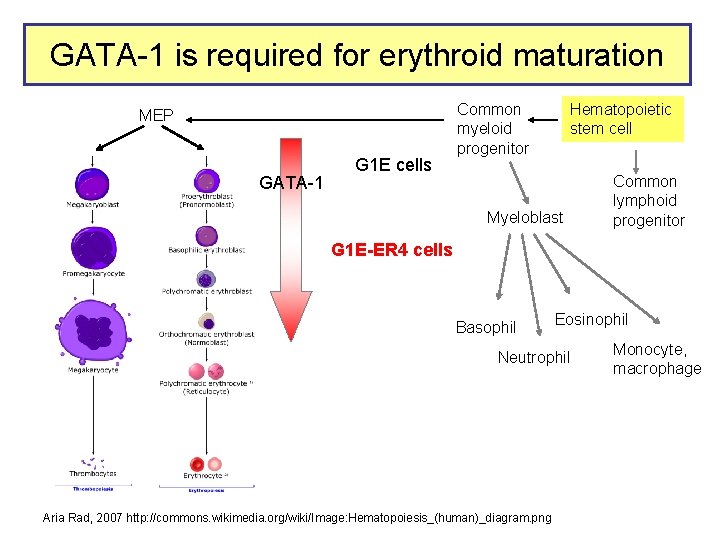
GATA-1 occupancy in erythroid cells

GATA-1 is required for erythroid maturation MEP GATA-1 G 1 E cells Common myeloid progenitor Hematopoietic stem cell Myeloblast Common lymphoid progenitor G 1 E-ER 4 cells Basophil Eosinophil Neutrophil Aria Rad, 2007 http: //commons. wikimedia. org/wiki/Image: Hematopoiesis_(human)_diagram. png Monocyte, macrophage

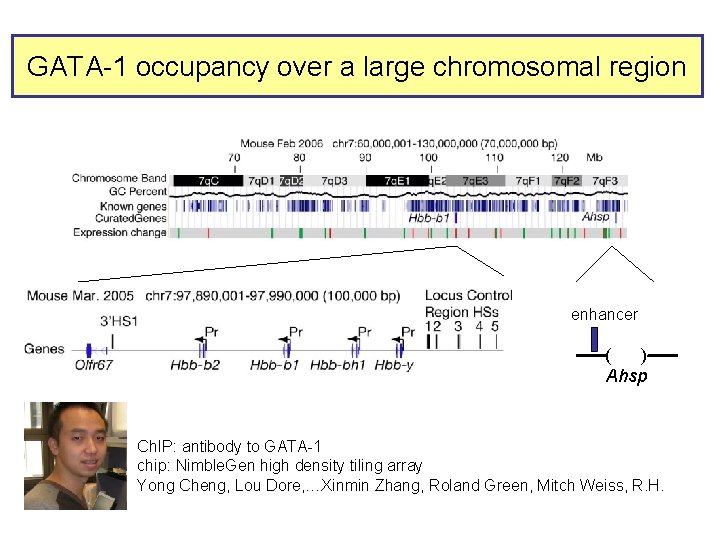
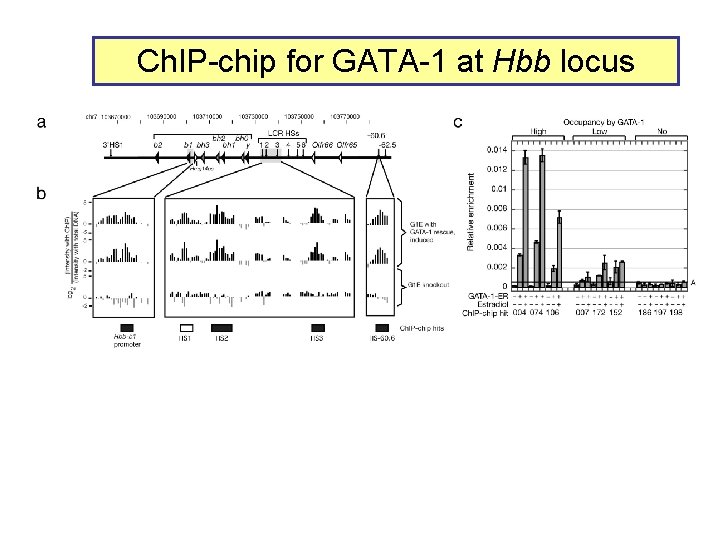
GATA-1 occupancy over a large chromosomal region enhancer ----( )---Ahsp Ch. IP: antibody to GATA-1 chip: Nimble. Gen high density tiling array Yong Cheng, Lou Dore, …Xinmin Zhang, Roland Green, Mitch Weiss, R. H.

Ch. IP-chip for GATA-1 at Hbb locus

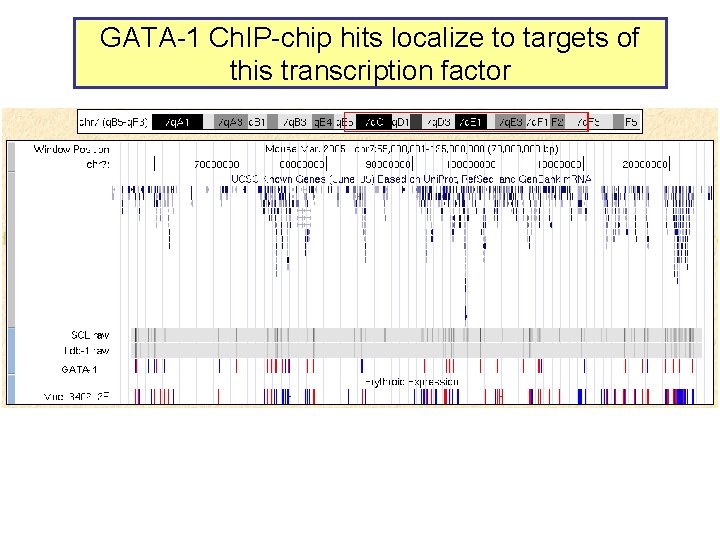
GATA-1 Ch. IP-chip hits localize to targets of this transcription factor GATA-1

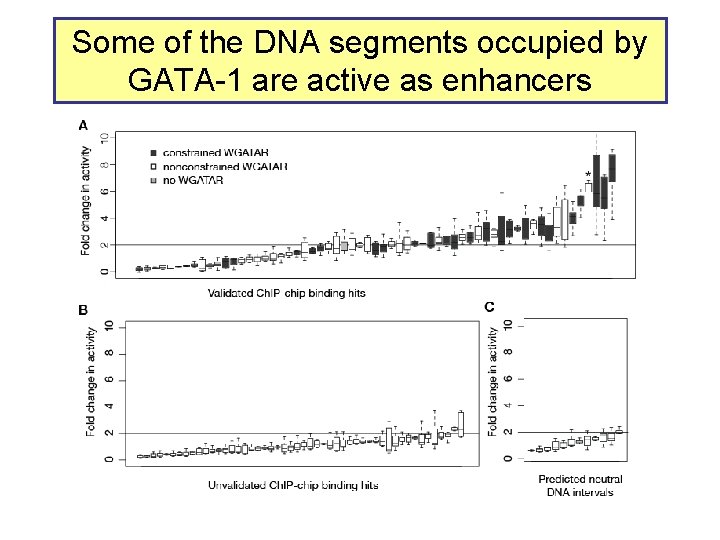
Almost all sites occupied by GATA-1 have the consensus binding site motif WGATAR • Of the 63 validated Ch. IP-chip hits, 60 (95%) have at least one WGATAR motif – Other 3 have AGATAT, GGATAT, CGATAG, … – Of 6000 randomly chosen DNA intervals of 500 bp from the 66 Mb, 3886 (65%) have a WGATAR motif – Occupied sites are about 1. 4 -fold enriched for the motif • GATA-1 discriminates exquisitely among available sites – Only 94 out of 78, 013 potential sites (500 bp interval with at least one WGATAR) are occupied – About 1 in 1000 intervals are occupied – Indicates exquisite specificity of the Ch. IP-chip data (<99%)

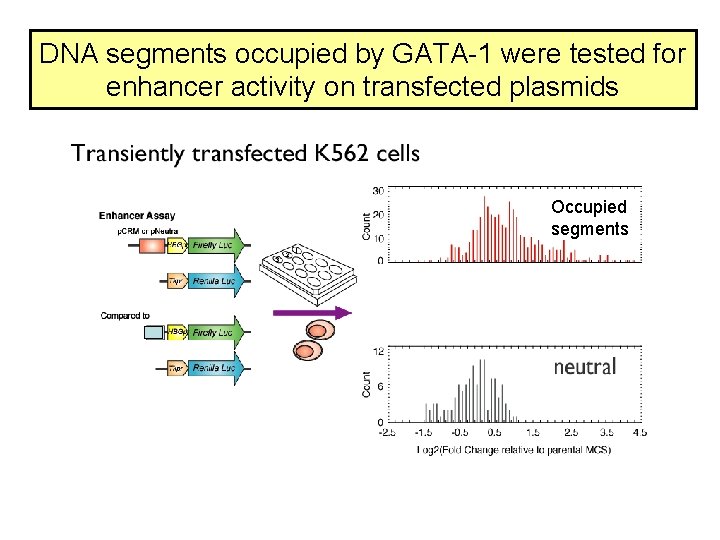
DNA segments occupied by GATA-1 were tested for enhancer activity on transfected plasmids Occupied segments

Some of the DNA segments occupied by GATA-1 are active as enhancers

Comparative genomics for predicting CRMs • Sometimes high quality data on biochemical signatures of CRMs is not available • Use sequence properties of CRMs for prediction • Clusters of binding site motifs for transcription factors – Low specificity - MANY false positives • Deep conservation of noncoding DNA sequences, from humans to fish or chicken – Low sensitivity - less than 5% of CRMs show signs of constraint across vertebrates • Conservation of clusters of transcription factor binding sites in mammals • Conservation patterns that distinguish CRMs from neutral DNA

Finding clusters of binding sites for transcription factors • Resources and servers for finding transcription factor binding sites (TFBSs) – – – TRANSFAC http: //www. gene-regulation. com/ JASPAR http: //jaspar. cgb. ki. se/cgi-bin/jaspar_db. pl TESS http: //www. cbil. upenn. edu/cgi-bin/tess MOTIF (Genome. Net) http: //motif. genome. jp/ Mat. Inspector http: //www. genomatix. de/

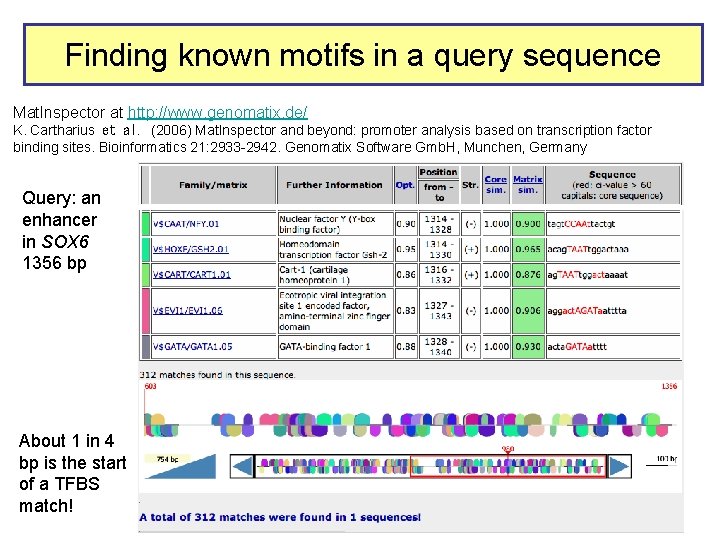
Finding known motifs in a query sequence Mat. Inspector at http: //www. genomatix. de/ K. Cartharius et al. (2006) Mat. Inspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933 -2942. Genomatix Software Gmb. H, Munchen, Germany Query: an enhancer in SOX 6 1356 bp About 1 in 4 bp is the start of a TFBS match!

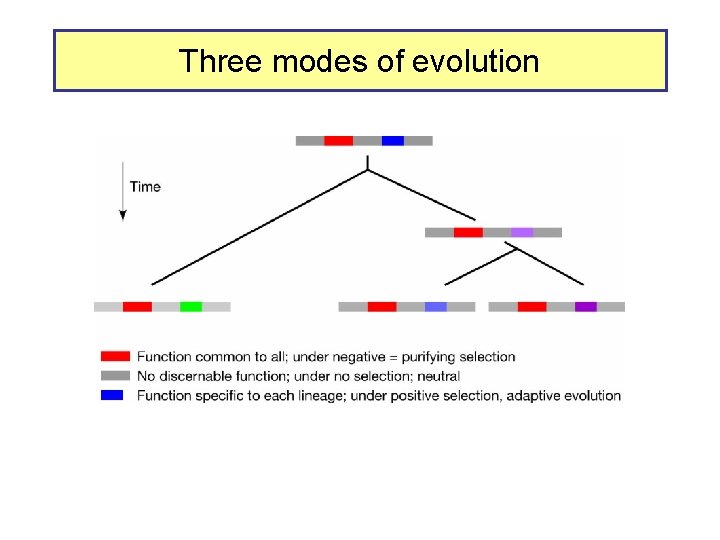
Three modes of evolution

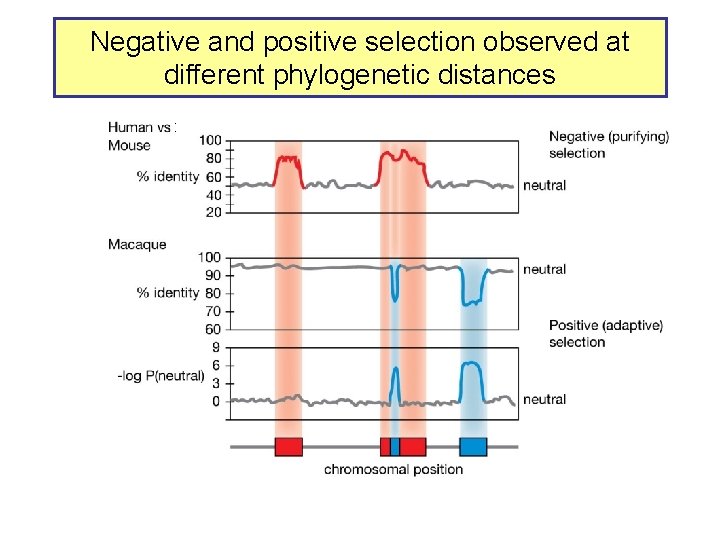
Negative and positive selection observed at different phylogenetic distances :

phast. Cons score identifies conserved DNA segments Siepel et al. 2005, Genome Research

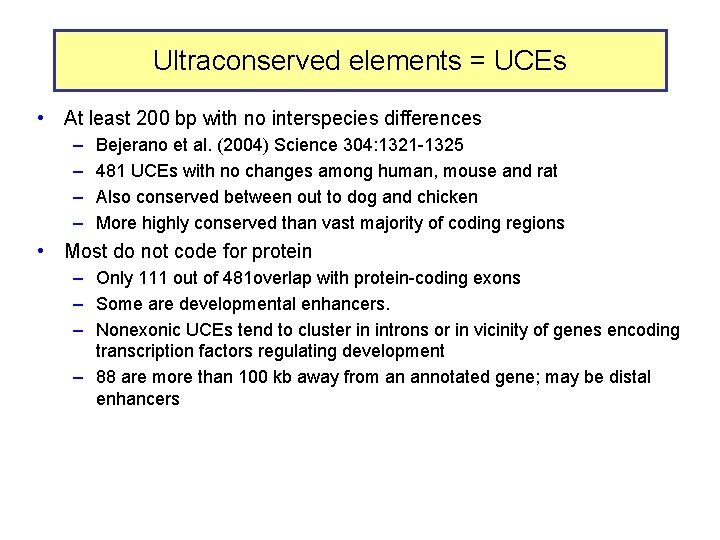
Ultraconserved elements = UCEs • At least 200 bp with no interspecies differences – – Bejerano et al. (2004) Science 304: 1321 -1325 481 UCEs with no changes among human, mouse and rat Also conserved between out to dog and chicken More highly conserved than vast majority of coding regions • Most do not code for protein – Only 111 out of 481 overlap with protein-coding exons – Some are developmental enhancers. – Nonexonic UCEs tend to cluster in introns or in vicinity of genes encoding transcription factors regulating development – 88 are more than 100 kb away from an annotated gene; may be distal enhancers

Intronic UCE in SOX 6 enhances expression in melanocytes in transgenic mice UCEs Tested UCEs Pennacchio et al. , http: //enhancer. lbl. gov/

Distinctive divergence rates for different types of functional DNA sequences p. TRRs: putative transcriptional regulatory region; likely CRMs Sites identified as occupied by sequence-specific transcription factors based on high-throughput chromatin immunoprecipitation assayed by hybridization to high density tiling arrays of genomic DNA= Ch. IP-chip

Genes likely regulated by clade-specific p. TRRs are enriched for distinctive functions Percentage of p. TRRs that align no further than: David King Primates: 3% Millions of years 91 Eutherians: 71% 173 310 Marsupials: 21% Enriched GO categories q-value for FDR Immune response 0. 0006 Protease inhibition Ion transport Mitosis and cell cycle 0. 0005 0. 012 0. 0005 450 Tetrapods: 4% Transcriptional 0. 004 regulation Vertebrates: 1% King, Taylor, et al. (2007) Genome Research

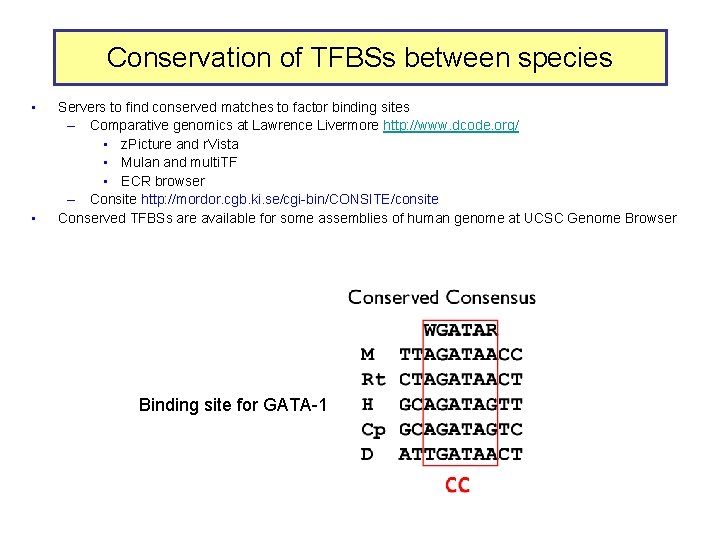
Conservation of TFBSs between species • • Servers to find conserved matches to factor binding sites – Comparative genomics at Lawrence Livermore http: //www. dcode. org/ • z. Picture and r. Vista • Mulan and multi. TF • ECR browser – Consite http: //mordor. cgb. ki. se/cgi-bin/CONSITE/consite Conserved TFBSs are available for some assemblies of human genome at UCSC Genome Browser Binding site for GATA-1

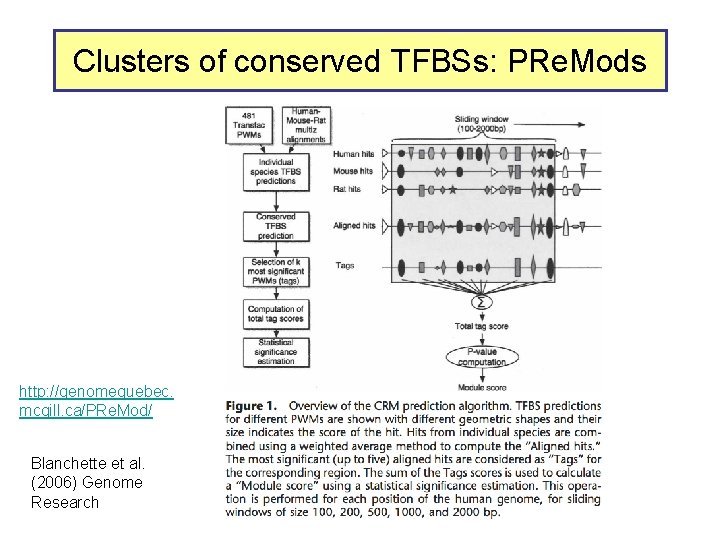
Clusters of conserved TFBSs: PRe. Mods http: //genomequebec. mcgill. ca/PRe. Mod/ Blanchette et al. (2006) Genome Research

ESPERR Evolutionary and Sequence Pattern Extraction through Reduced Representation Taylor et al. (2006) Genome Research 16: 1596 -1604

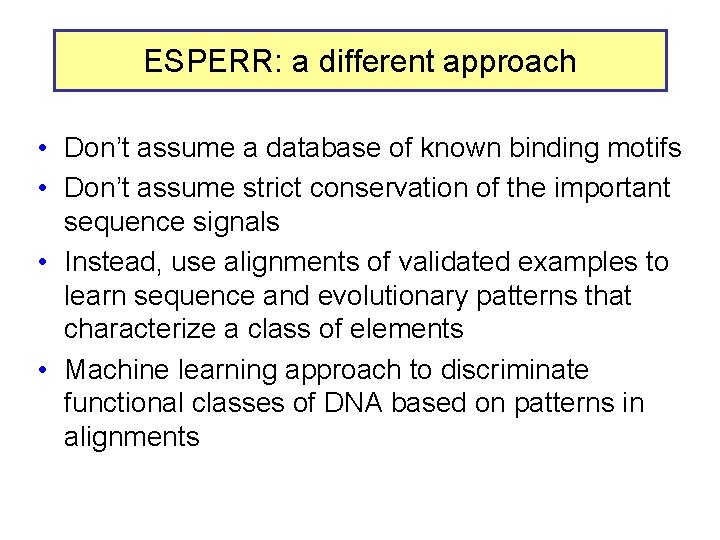
ESPERR: a different approach • Don’t assume a database of known binding motifs • Don’t assume strict conservation of the important sequence signals • Instead, use alignments of validated examples to learn sequence and evolutionary patterns that characterize a class of elements • Machine learning approach to discriminate functional classes of DNA based on patterns in alignments

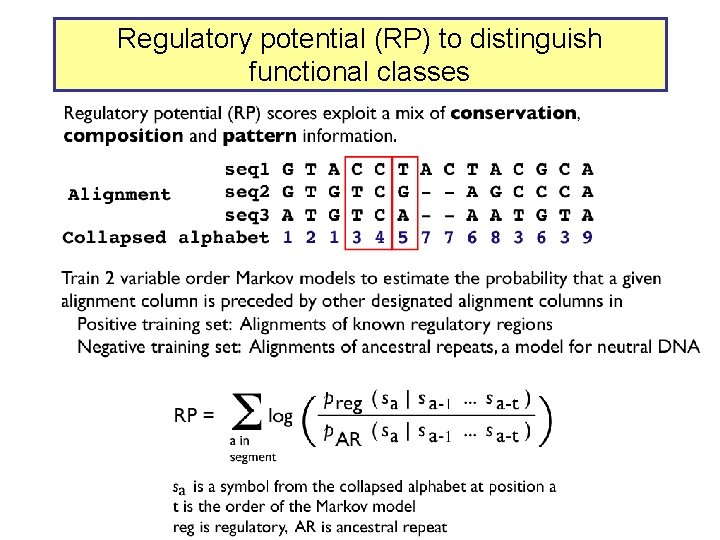
Regulatory potential (RP) to distinguish functional classes

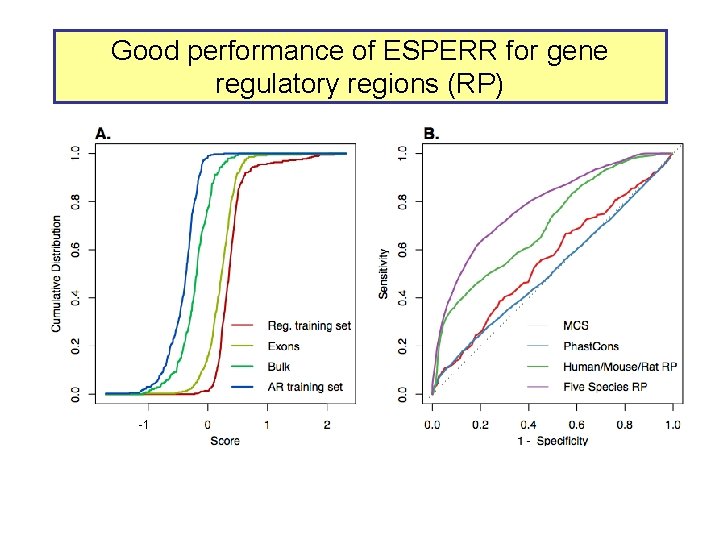
Good performance of ESPERR for gene regulatory regions (RP) -1

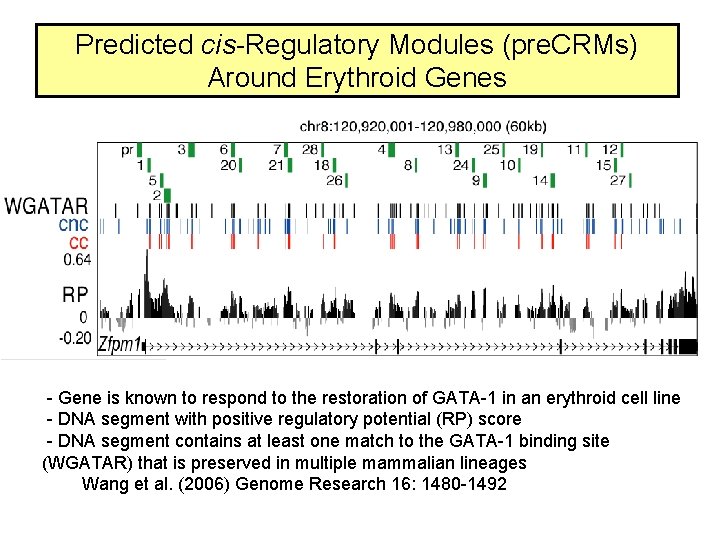
Predicted cis-Regulatory Modules (pre. CRMs) Around Erythroid Genes - Gene is known to respond to the restoration of GATA-1 in an erythroid cell line - DNA segment with positive regulatory potential (RP) score - DNA segment contains at least one match to the GATA-1 binding site (WGATAR) that is preserved in multiple mammalian lineages Wang et al. (2006) Genome Research 16: 1480 -1492

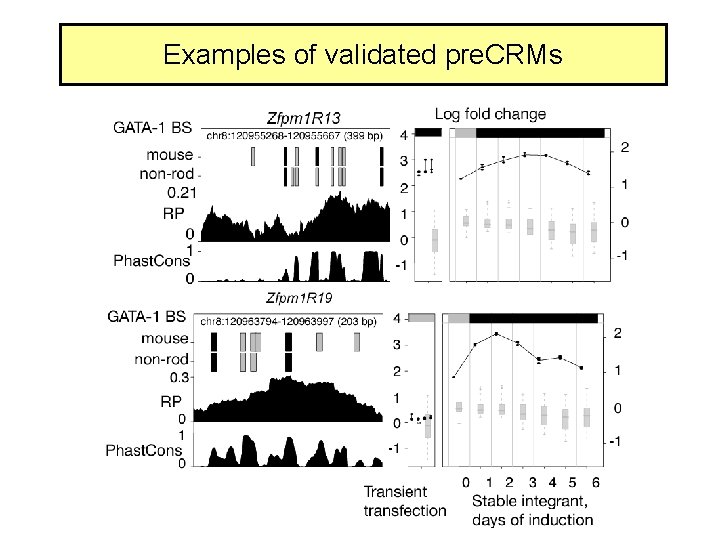
Examples of validated pre. CRMs

Validation status for 99 tested fragments cc = consensus binding site motif is conserved and matches the consensus in multiple mammalian lineages cnc = binding site motif has a mismatch from the consensus but is conserved Wang et al. (2006) Genome Research 16: 1480 -1492

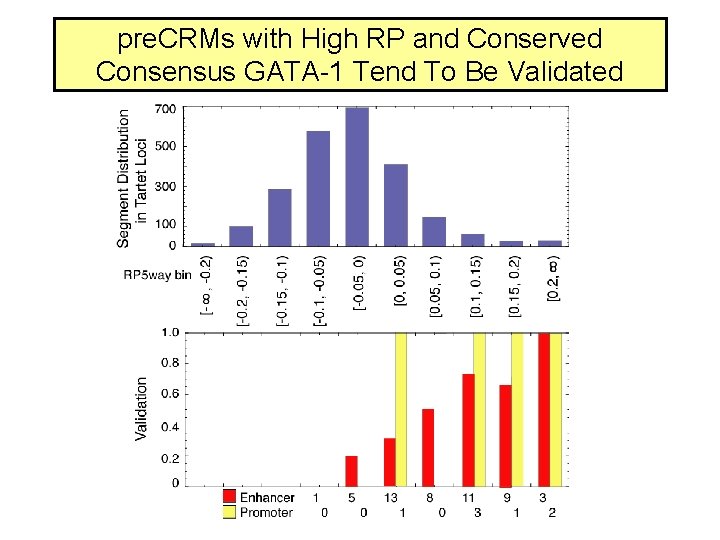
pre. CRMs with High RP and Conserved Consensus GATA-1 Tend To Be Validated

Accurate prediction of a GATA-1 responsive enhancer for mi. R-144, 451 A Dore L, Amigo JD et al. (2008) PNAS 105: 3333 -3338.

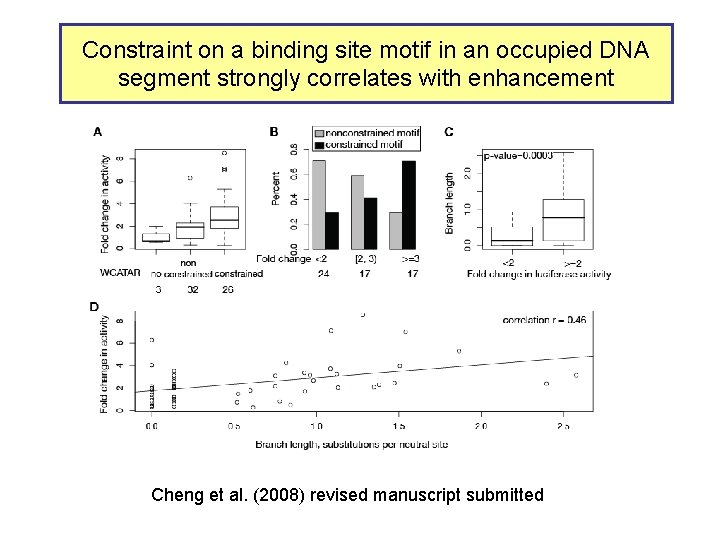
Constraint on a binding site motif in an occupied DNA segment strongly correlates with enhancement Cheng et al. (2008) revised manuscript submitted

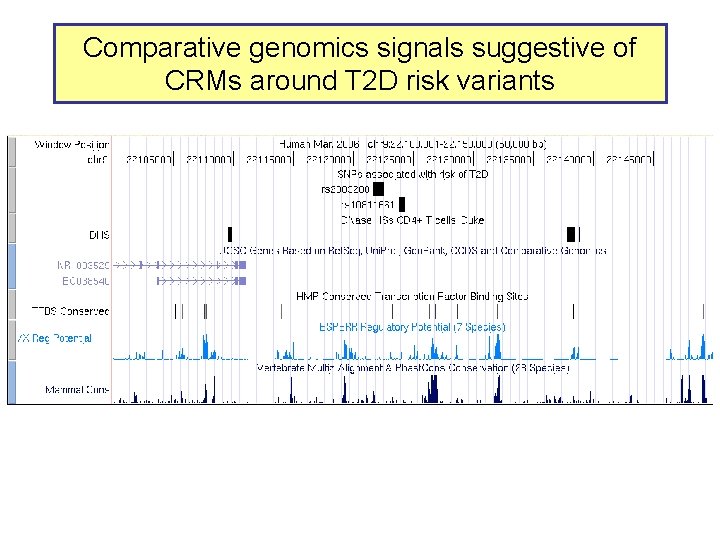
Comparative genomics signals suggestive of CRMs around T 2 D risk variants

Summary: Genomics of Gene Regulation • • • Genetic determinants of variation in expression levels may contribute to complex traits - phenotype is not just determined by coding regions Biochemical features associated with cis-regulatory modules are being determined genome-wide for a range of cell types. These can be used to predict CRMs, but occupancy does not necessarily mean that the DNA is actively involved in regulation. Comparative genomics is a complementary approach to predicting CRMs. Evolutionary preservation of binding site motifs within regions containing other indicators of CRMs (e. g. regulatory potential or protein occupancy) is a good predictor of function.

Many thanks … RP scores and other bioinformatic input: Francesca Chiaromonte, James Taylor Yong Cheng, Demesew Abebe, Christine Dorman, …, Ying Zhang, David King, Swathi Ashok Kumar Erythroid cell biology and biochemistry: Mitch Weiss, Gerd Blobel, Barry Paw Alignments, chains, nets, browsers, ideas, … Webb Miller, Jim Kent, David Haussler Funding from NIDDK, NHGRI, Huck Institutes of Life Sciences at PSU