Genomics Genetic Analysis on a Genomewide global scale
Genomics • Genetic Analysis on a Genome-wide (global) scale – Answers to questions about development, disease, evolution • Whole Genome Mapping – High Resolution Genetic Maps • High resolution meiotic recombination maps • High resolution cytogenetic maps – Physical Genomic Maps • “Ordered clones” or “Contigs” – Sequence Maps (highest possible resolution) • Bioinformatics – Computational Analysis of Sequence information • DNA and Protein • Functional Genomics – Experimental Analysis to determine the function of all genome components
High resolution genetic (meiotic recombination) maps • What good are Genome Maps? – Provide markers for finding location of genes (disease/developmental genes) – Provide markers for assembling individual clones into whole chromosomes • Why is high resolution necessary? – Cytological or recombinational maps get you to within 1 Mb of gene • High Reolustion means high density of Genetic Markers – Genetic Markers = Polymorphisms Used in Mapping • Genes • Neutral DNA sequence variations (DNA polymorphisms) • DNA polymorphisms – RFLP (restriction fragment length polymorphisms) – Simple Sequence Length Polymorphisms (SSLPs) • Minisatellite sequences (aka VNTR = variable number of tandem repeats) • Microsatellite sequences (aka STRs =Simple Tandom Repeats) • RAPD (Randomly Amplified Polymorphic DNA) – SNPs (Single Nucleotide Polymorphisms) • Single nucleotide differences in the sequence of chromosomes – between homologs; between individuals; high heterozygosity
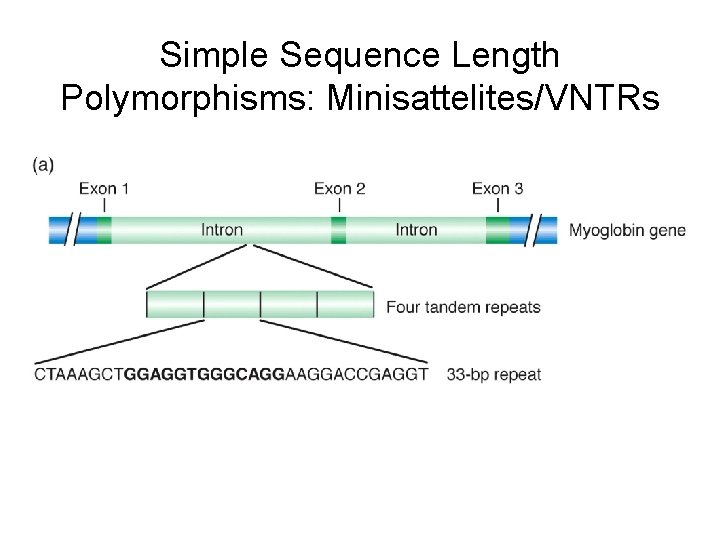
Simple Sequence Length Polymorphisms: Minisattelites/VNTRs
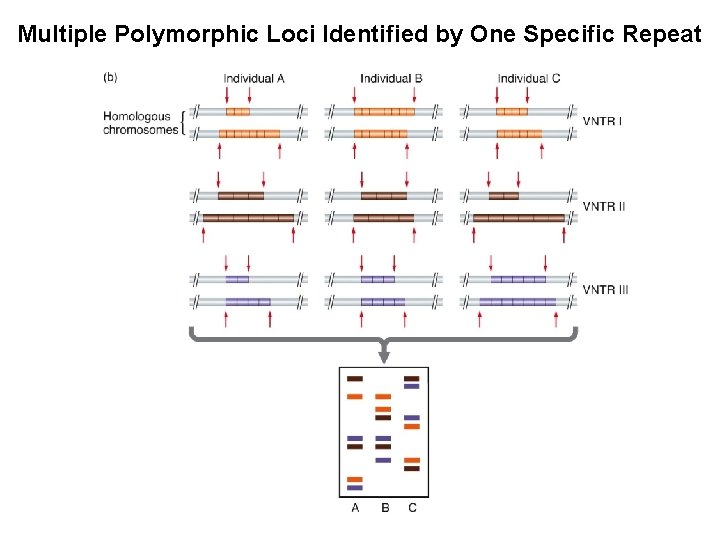
Multiple Polymorphic Loci Identified by One Specific Repeat
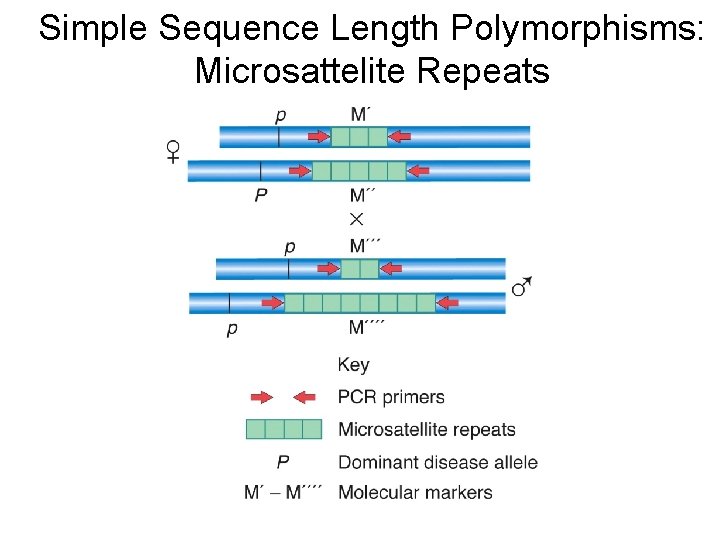
Simple Sequence Length Polymorphisms: Microsattelite Repeats
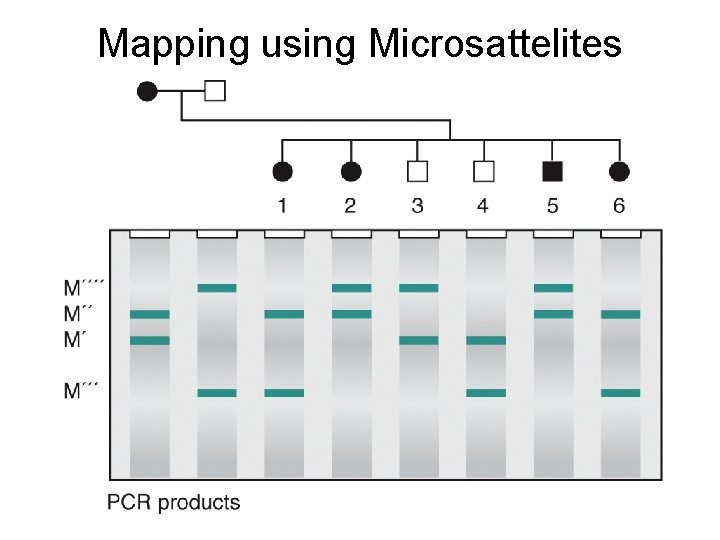
Mapping using Microsattelites
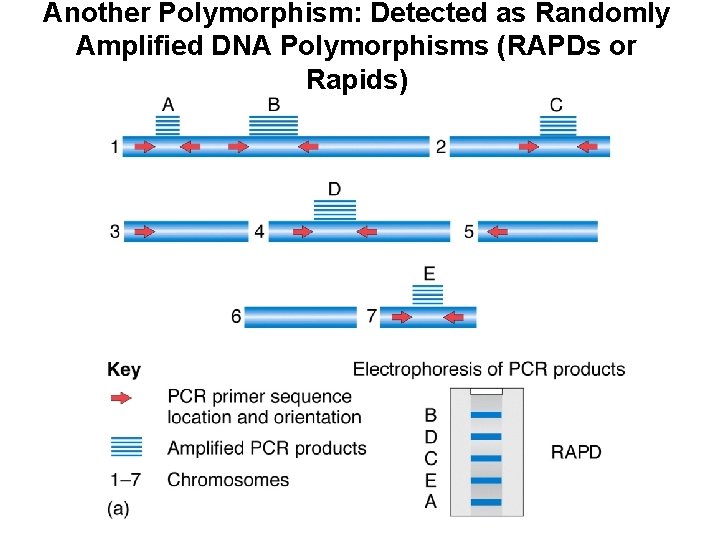
Another Polymorphism: Detected as Randomly Amplified DNA Polymorphisms (RAPDs or Rapids)
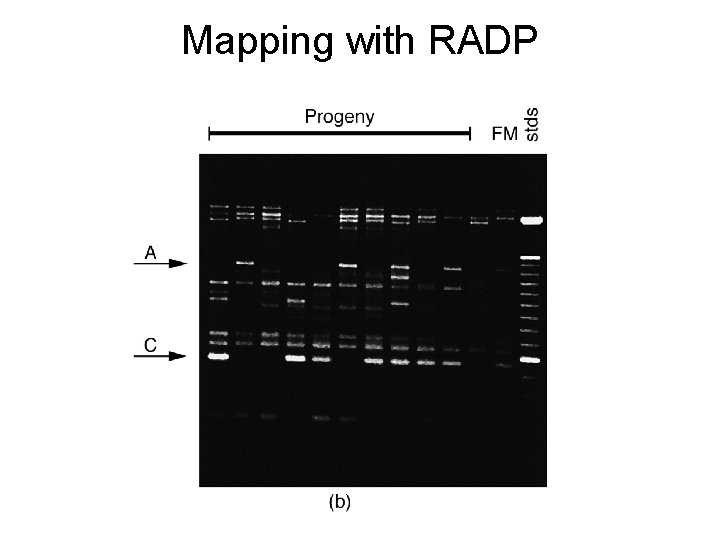
Mapping with RADP
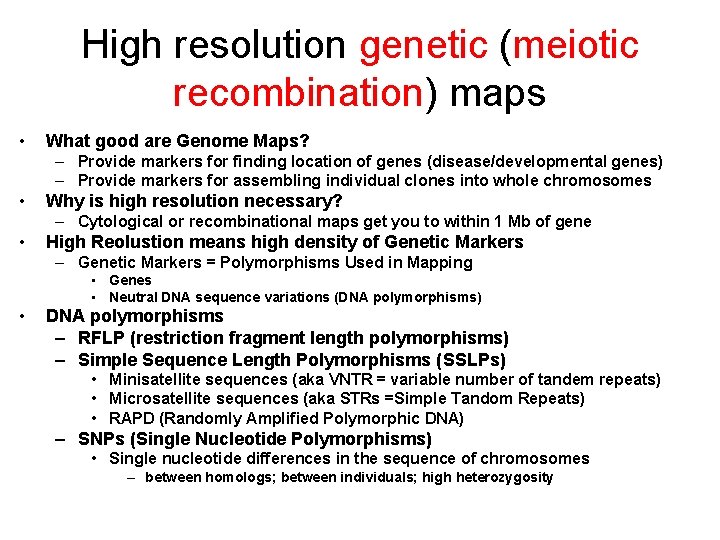
High resolution genetic (meiotic recombination) maps • What good are Genome Maps? – Provide markers for finding location of genes (disease/developmental genes) – Provide markers for assembling individual clones into whole chromosomes • Why is high resolution necessary? – Cytological or recombinational maps get you to within 1 Mb of gene • High Reolustion means high density of Genetic Markers – Genetic Markers = Polymorphisms Used in Mapping • Genes • Neutral DNA sequence variations (DNA polymorphisms) • DNA polymorphisms – RFLP (restriction fragment length polymorphisms) – Simple Sequence Length Polymorphisms (SSLPs) • Minisatellite sequences (aka VNTR = variable number of tandem repeats) • Microsatellite sequences (aka STRs =Simple Tandom Repeats) • RAPD (Randomly Amplified Polymorphic DNA) – SNPs (Single Nucleotide Polymorphisms) • Single nucleotide differences in the sequence of chromosomes – between homologs; between individuals; high heterozygosity
High Resolution Cytogenetic maps
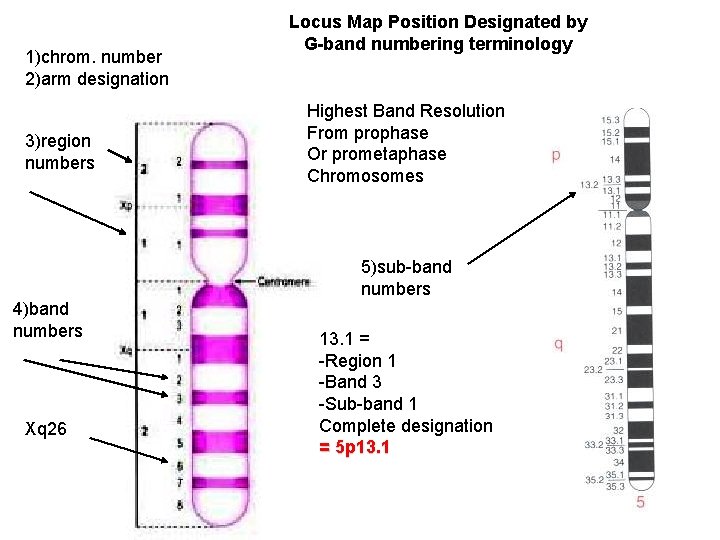
1)chrom. number 2)arm designation 3)region numbers Locus Map Position Designated by G-band numbering terminology Highest Band Resolution From prophase Or prometaphase Chromosomes 5)sub-band numbers 4)band numbers Xq 26 13. 1 = -Region 1 -Band 3 -Sub-band 1 Complete designation = 5 p 13. 1

In Situ Hybridization Mapping
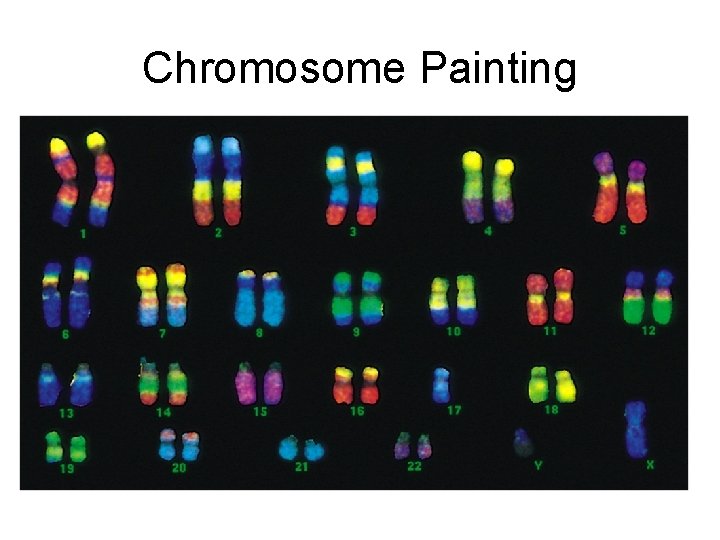
Chromosome Painting
Genomics • Genetic Analysis on a Genome-wide (global) scale – Answers to questions about development, disease, evolution • Whole Genome Mapping – High Resolution Genetic Maps • High resolution meiotic recombination maps • High resolution cytogenetic maps – In situ Hybridization » On G banded or “Painted” chromosomes – Rearrangement Breakpoint Mapping – Radiation Hybrid Mapping – Physical Genomic Maps • “Ordered clones” or “Contigs” – Ordering by fingerprinting – Sequence Maps (highest possible resolution) • Bioinformatics – Computational Analysis of Sequence information • DNA and Protein • Functional Genomics – Experimental Analysis to determine the function of all genome components
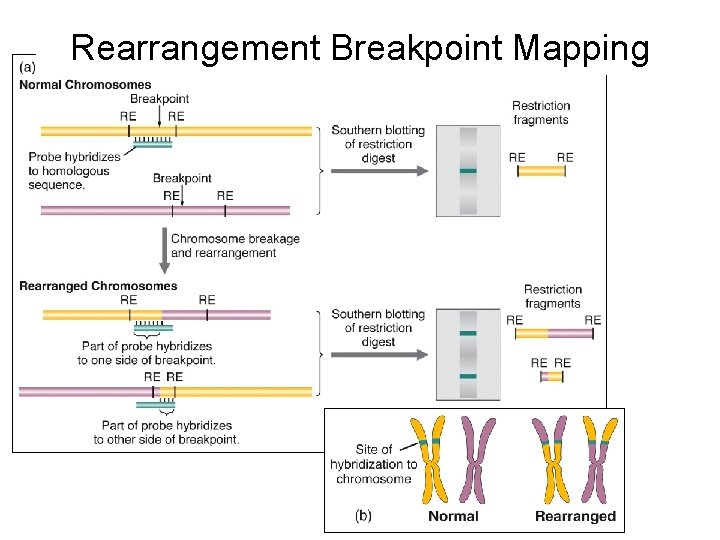
Rearrangement Breakpoint Mapping
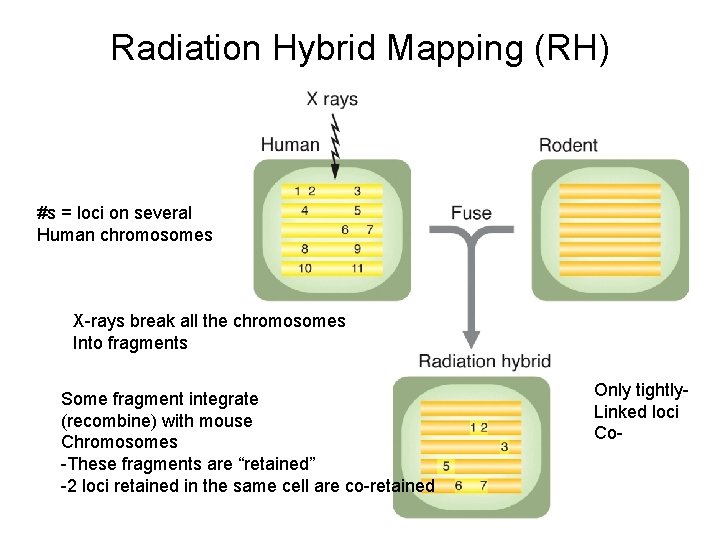
Radiation Hybrid Mapping (RH) #s = loci on several Human chromosomes X-rays break all the chromosomes Into fragments Some fragment integrate (recombine) with mouse Chromosomes -These fragments are “retained” -2 loci retained in the same cell are co-retained Only tightly. Linked loci Co-
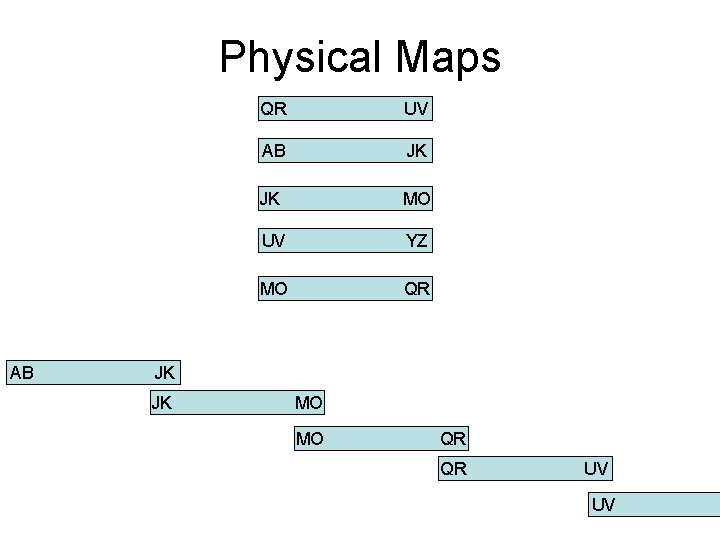
Physical Maps AB QR UV AB JK JK MO UV YZ MO QR JK JK MO MO QR QR UV UV
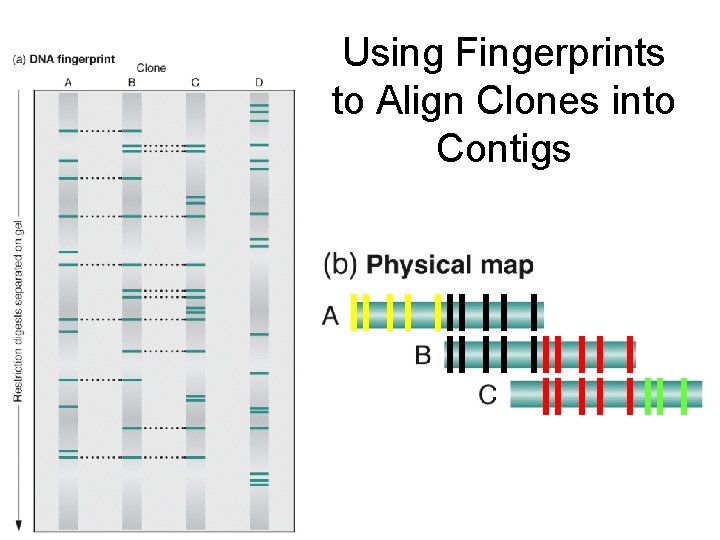
Using Fingerprints to Align Clones into Contigs
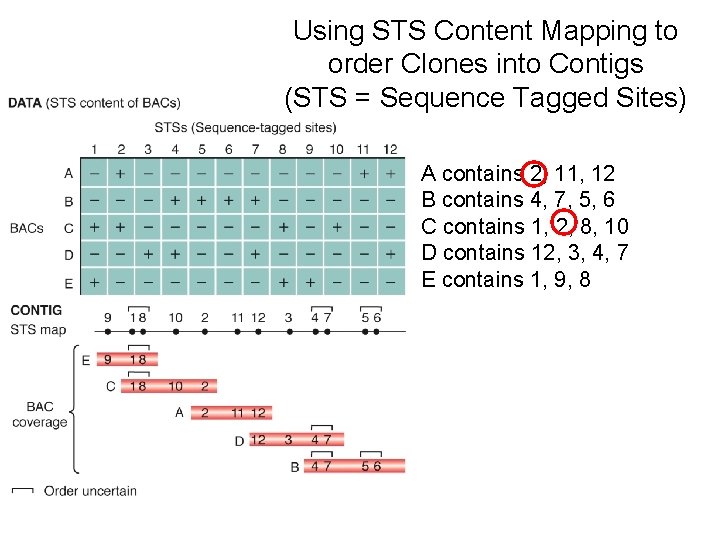
Using STS Content Mapping to order Clones into Contigs (STS = Sequence Tagged Sites) A contains 2, 11, 12 B contains 4, 7, 5, 6 C contains 1, 2, 8, 10 D contains 12, 3, 4, 7 E contains 1, 9, 8
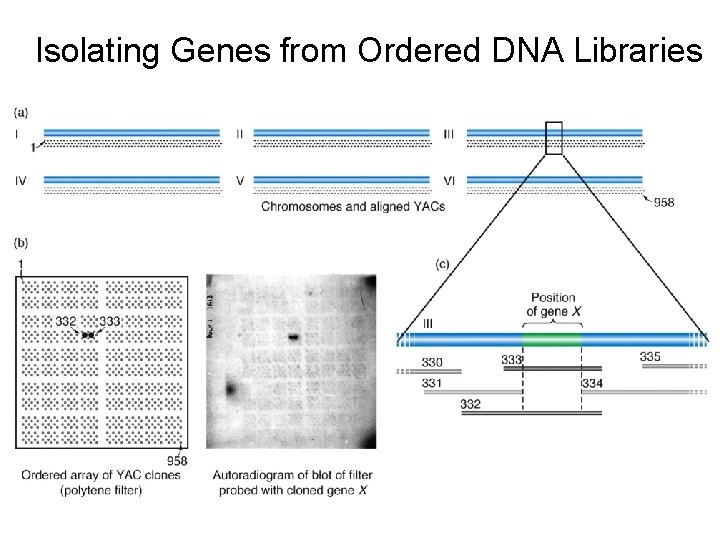
Isolating Genes from Ordered DNA Libraries

Subdividing the Genome • Will not cover this • Has to do with making libraries of individual chromosomes or fragments of chromosomes instead of the whole genome • Gets around some biological and technical limitations of the technology

Sequencing the Genome • • • Consensus Sequence Assembly Ordered Clone Sequencing Whole Genome Shotgun Sequencing Sequence Contig Assembly Problems Caused by Repetitive DNA Tackling Problems caused by Repetitive DNA

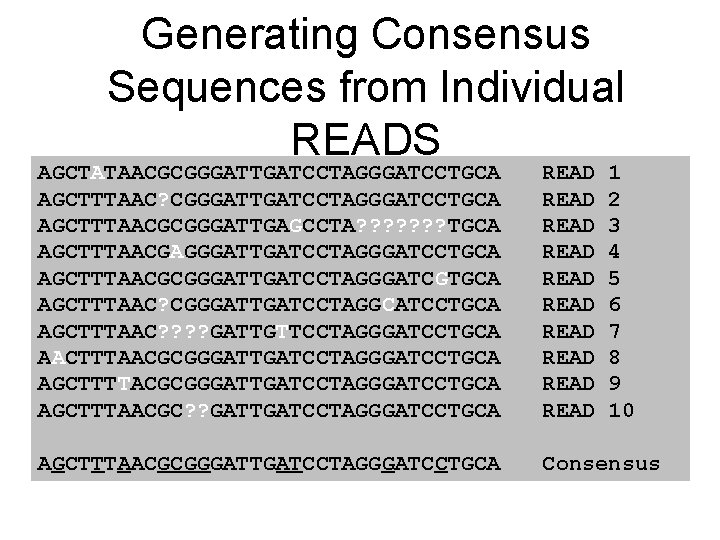
Generating Consensus Sequences from Individual READS AGCTATAACGCGGGATTGATCCTAGGGATCCTGCA AGCTTTAAC? CGGGATTGATCCTAGGGATCCTGCA AGCTTTAACGCGGGATTGAGCCTA? ? ? ? TGCA AGCTTTAACGAGGGATTGATCCTAGGGATCCTGCA AGCTTTAACGCGGGATTGATCCTAGGGATCGTGCA AGCTTTAAC? CGGGATTGATCCTAGGCATCCTGCA AGCTTTAAC? ? GATTGTTCCTAGGGATCCTGCA AACTTTAACGCGGGATTGATCCTAGGGATCCTGCA AGCTTTTACGCGGGATTGATCCTAGGGATCCTGCA AGCTTTAACGC? ? GATTGATCCTAGGGATCCTGCA READ READ READ 1 2 3 4 5 6 7 8 9 10 AGCTTTAACGCGGGATTGATCCTAGGGATCCTGCA Consensus
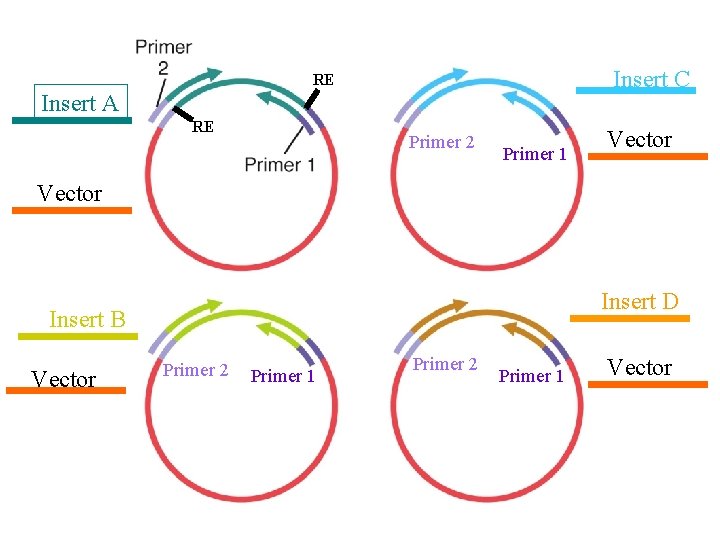
Insert C RE Insert A RE Primer 2 Primer 1 Vector Insert D Insert B Vector Primer 2 Primer 1 Vector
- Slides: 25