Genomics England update 1 Overview 1 Sample identity

















- Slides: 58

Genomics England update 1

Overview 1. Sample identity checks – Ellen Thomas 2. CIP update – Liz Edwards 3. Interpretation portal and tiering browser – Chris Boustred 4. Results flow and roadmap – Ellen Thomas+1 5. Validation update – Emma Baple 6. Q and A – All 06 May 2016 2

Where are we aiming to get to? 3

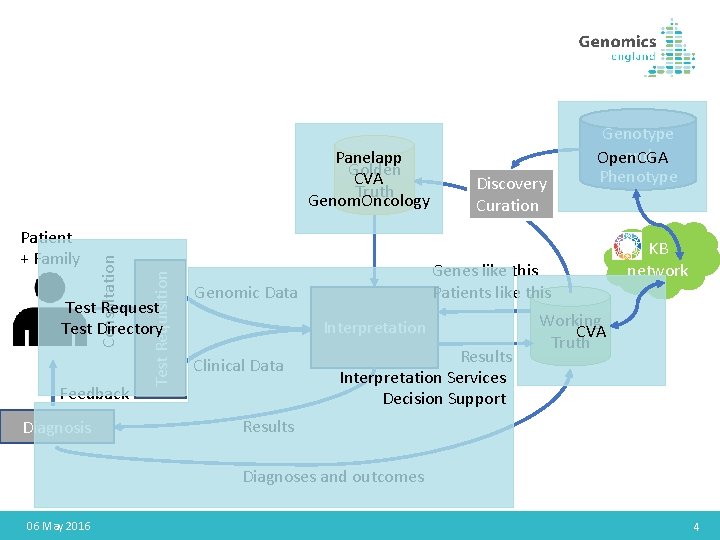
Test Requisition Patient + Family Consultation Panelapp Golden CVA Truth Genom. Oncology Test Request Test Directory Feedback Diagnosis Discovery Curation Genes like this Patients like this Genomic Data Interpretation Clinical Data Genotype and Open. CGA Phenotype Results Interpretation Services Decision Support KB network Working CVA Truth Results Diagnoses and outcomes 06 May 2016 4

Sample identity checks 5

Background • All samples undergo bioinformatics checks to determine: • Whether the genetic sex is the same as the sex stated in the clinical data • Whether relationships in the family are compatible with the pedigree supplied in the clinical data • Samples may fail these checks for different reasons: • • Data error in clinical data Sample swap Misattributed relationship Complex genetic mechanisms e. g. UPD 6

Extent of sample failures • In main programme checks carried out up to the end of 2017 (>5000 genomes): • 115 families affected by fails (majority borderline and required no action) • 44 of these required further investigation • 20 families affected by a deviation in data transfer for a single plate at Illumina in 2016: all samples blocked, none returned to NHS GMCs, investigation has indicated the data error which is likely to be remediable internally • 3 likely to represent other sample swaps (within or between families) • Remainder likely to be mainly data errors 7

Response to samples which fail checks • Clinical and bioinformatics teams review data together • Likely cause identified where possible • To date, these families have not started to be returned to NHS GMCs (except 2 cases of extreme urgency where a solution was agreed with the NHS GMC) • Proposed NHS GMC-facing SOP now developed • Individual query lists to be returned to NHS GMCs after that (detailed information supplied via nhs. net to responsible clinician) 8

NHS GMC responsibilities • New sample can be provided where relevant • Data errors can be corrected by resubmitting corrected information • Individual samples can be removed from the analysis leaving the remaining relative(s) to proceed through the pipeline where this is the best outcome for that family 9

Interpretation portal and tiering browser 10

Outline • Case browser • Performance • New filters • Preserving filter settings • “Interpretation” Browser • • Exomiser results Search variants Save work in progress Pub. Med search • Exit Questionnaire • Save work in progress • Other • Link to service desk • Panel names colour coding • Various bugfixes November 2020 11

Case Browser

Performance • Currently: ~35 seconds to load “To Be Reviewed” tab in production • Initial tests: ~2. 3 seconds • 16 x improvement November 2020 13

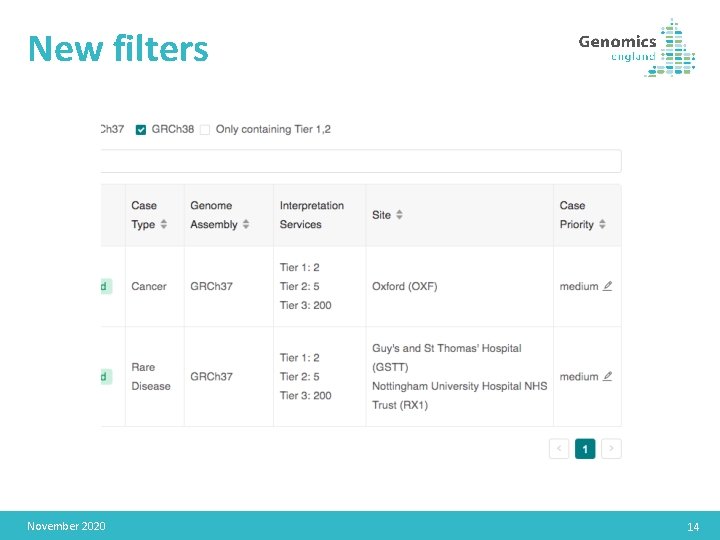
New filters November 2020 14

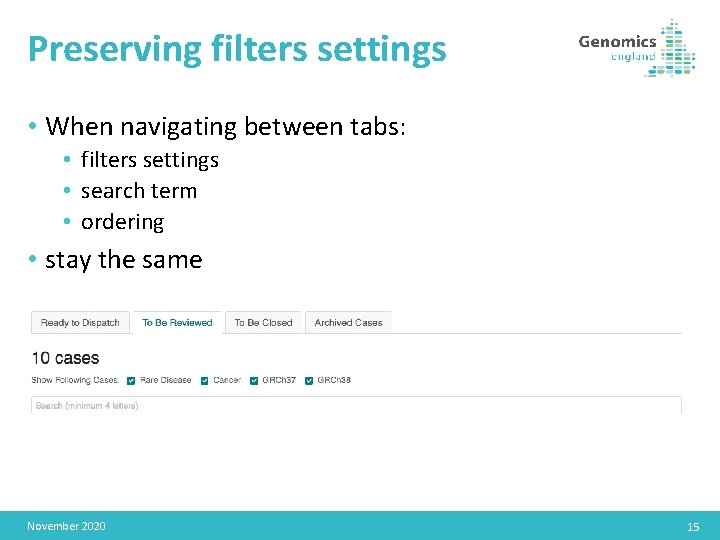
Preserving filters settings • When navigating between tabs: • filters settings • search term • ordering • stay the same November 2020 15

Interpretation Browser

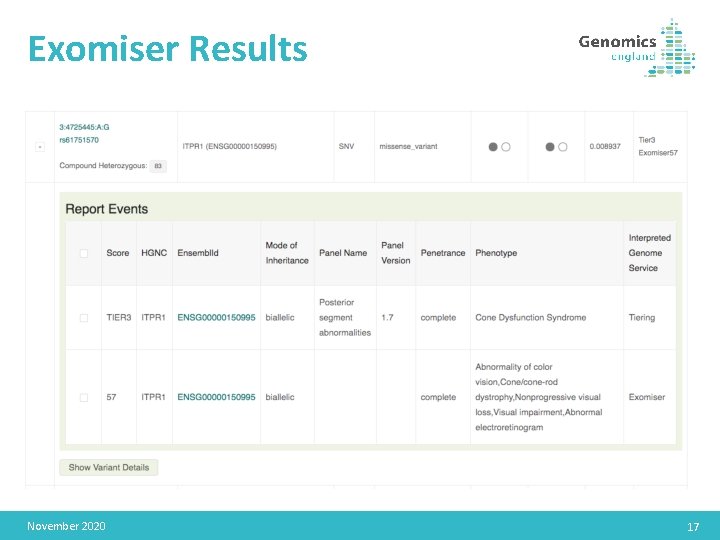
Exomiser Results November 2020 17

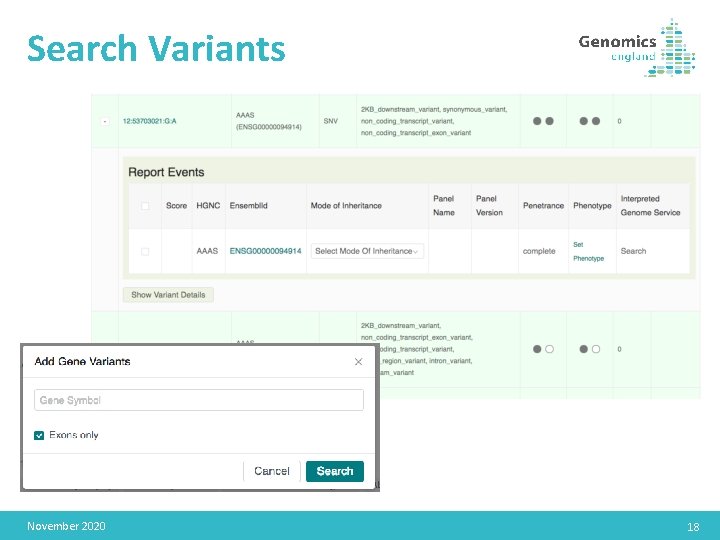
Search Variants November 2020 18

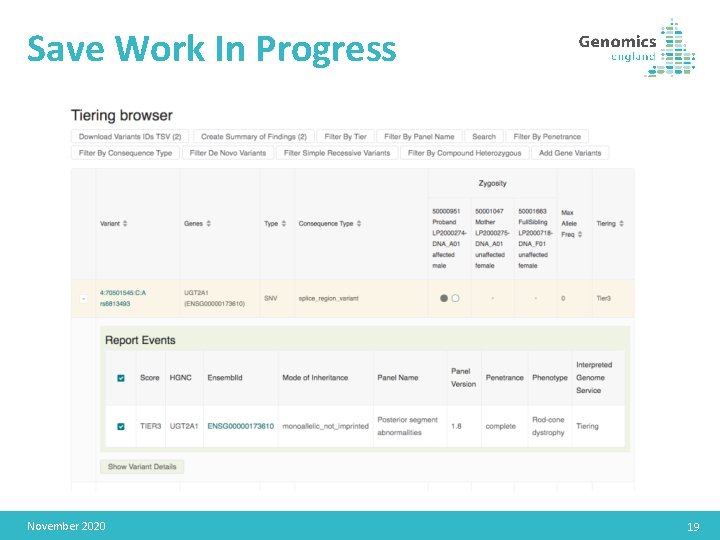
Save Work In Progress November 2020 19

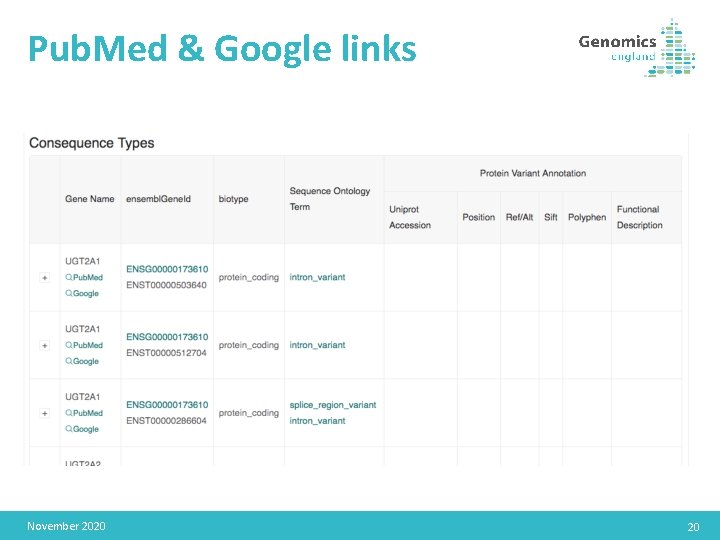
Pub. Med & Google links November 2020 20

Exit Questionnaire

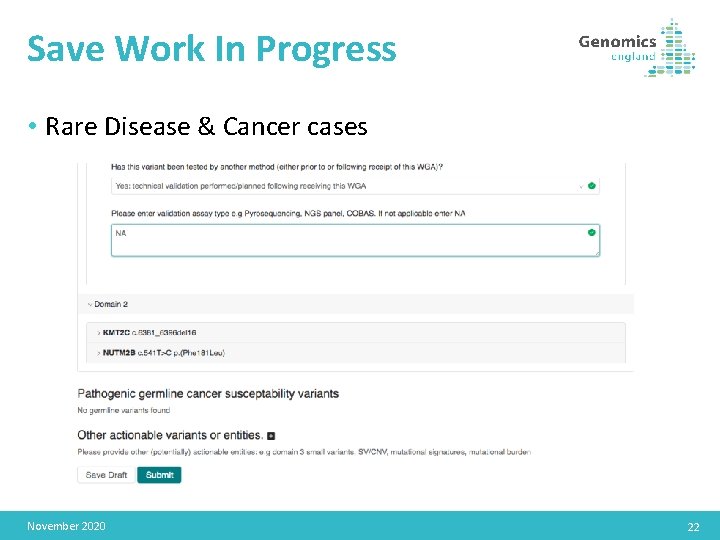
Save Work In Progress • Rare Disease & Cancer cases November 2020 22

Other

Panel Names Colour Coding November 2020 24

Link to Service Desk November 2020 25

GMC Feedback (non exhaustive list) • • Filter cases with Tier 1 and Tier 2 variants Assign cases to users – approving / checking More variant quality information e. g. ref / alt depth Ability to: • Apply new panel • “Re-tier” • • • To be internally reviewed and prioritised Batch complete Outcomes Questionnaires Display CNVs IGV performance HGVS Additional links – OMIM, HGMD, Clin. Var etc. ACMG classification Previous interpretations / classifications Ref Seq transcripts Link to Documentation Splice prediction tools … November 2020 26

Multiple monogenic conditions • Families recruited who may have multiple monogenic conditions which don’t segregate together • E. g. child with intellectual disability, father with cardiomyopathy • Tiering can be run several times per family, using the disorders attached to each individual to allocate segregation patterns to particular panels • Pipeline developments to allow this currently in test • CIPs aiming to develop methods to display these; will be returned in the tiering browser in any case 06 May 2016 27

CIP update 28

CIP grch 38 Deployment • Congenica: • Deployed Sapientia v 1. 8 with grch 38 support 16 th April 2018 • Initial dispatch approach has been to dispatch a small batch of cases to each NHS GMC to ensure NHS GMC workflows are suitably aligned. • So far 120 grch 38 cases successfully dispatched to NHS GMCs. • This week cases will begin to be returned in earnest, to clear the backlog of unclosed grch 38 cases in the Interpretation Portal and keep pace with new cases entering the system. • Fabric: • Opal with grch 38 support is planned to launch week commencing 14 th May 2018. • Initial dispatch strategy will be similar to that of Congenica to ensure compatibility with NHS GMC processes. November 2020 29

CIPs of the Future: CIP Tender • Initial CIP contracts were for 2000 cases, this has been extended until the CIP Tender has completed. • Due to contract values, a formal tendering process is required to cover the remainder of the 100 KGP. • As part of the tender process a detailed specification was created with Genomics England Clinical Team and significant input from a group of NHS GMC volunteers across 5 NHS GMCs. • 5 Tenderers were shortlisted, with 1 dropping out prior to the interview stage. • Congenica, Fabric, Agilent and Qiagen were the final shortlisted Decision Support Providers. Interviews and a technical challenge were conducted last week. November 2020 30

CIPs of the Future: CIP Tender • Genomics England tender evaluation team: • • • Nick Maltby (General Counsel & Company Secretary) Augusto Rendon (Director of Bioinformatics) Paul Nicholson (Procurement Lead) Christopher Boustred (Clinical Bioinformatics Lead) Liz Edwards (Head of Bioinformatics Partnerships) Olivia Niblock (Clinical Bioinformatics Analyst) • Final evaluation scheduled for 15 th May 2018 • Commencement of services: 19 th July 2018 November 2020 31

CIPs of the Future: Illumina BSVI • In addition to the CIP(s) that win the CIP tender, Illumina have, in partnership with Genomics England, been developing their Base. Space Variant Interpreter (BSVI) Platform. • Having multiple CIP providers ensures there is redundancy in the system and capacity for throughput. • BSVI is a decision support tool for both Cancer and Rare Disease. • BSVI Cancer offering is already being rolled out to NHS GMCs since Dec 2017. • BSVI Rare Disease platform scheduled for a pilot roll out to NHS GMCs from July 2018. November 2020 32

CIPS of the Future: Illumina BSVI • BSVI Rare Disease platform has been developed towards the CIP Tender specification. • All open cases and cases ready for interpretation in the system will be available in BSVI Rare Disease. • Details of the BSVI Rare Disease pilot phase roll out process will be distributed to NHS GMCs soon. November 2020 33

Results flow and roadmap 34

Background • Although results from the main programme have begun to be returned to NHS GMCs over the last 18 months, the rate of return and the overall volumes have differed greatly between each site • The systems and processes for the return of results have required extensive iteration and development • As we look ahead we need to agree a strategic approach to manage the remaining (majority of) results from the Project that Genomics England plan to release by the end of March 2019 and consider how re-analysis is incorporated November 2020 35

Aim The purpose of this session is to help create a consensus between the NHS GMCs, NHS England Genomics England, about our approach to handling the return of the results to support and enable each site to plan effectively. November 2020 36

Key areas of focus There are different challenges that need to be addressed across the different elements of the results pipeline. To help develop an approach, we have considered the challenges across the following elements: 1. Rare disease main findings (build 37) 2. Rare disease main findings (backlog on build 38) 3. Rare disease main findings (prospective, both builds) 4. Additional findings 5. Reanalysis to incorporate improvements in the pipeline • • Realignment to build 38 CNVs/SVs Tiering whitelist Improved gene panels November 2020 37

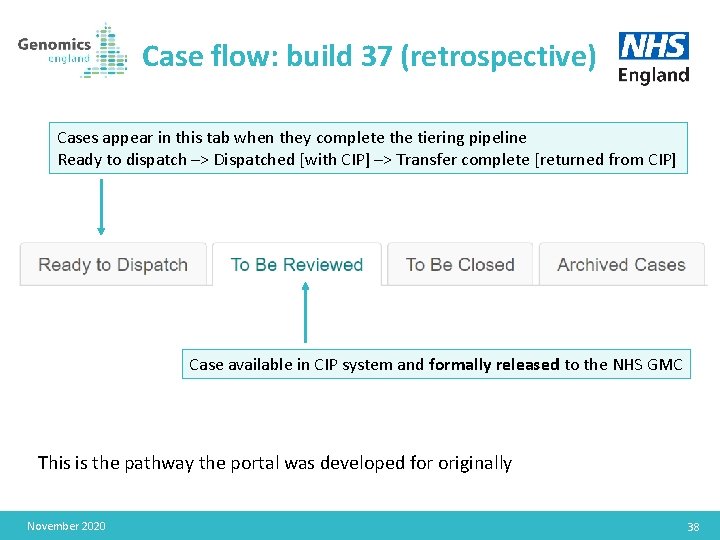
Case flow: build 37 (retrospective) Cases appear in this tab when they complete the tiering pipeline Ready to dispatch –> Dispatched [with CIP] –> Transfer complete [returned from CIP] Case available in CIP system and formally released to the NHS GMC This is the pathway the portal was developed for originally November 2020 38

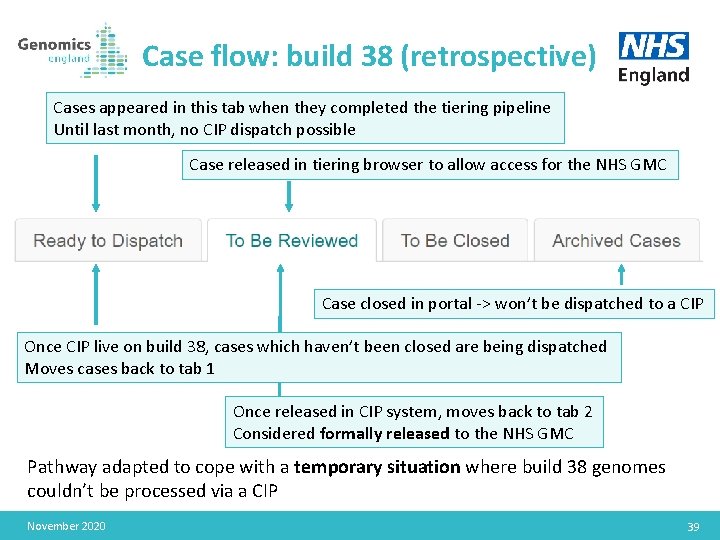
Case flow: build 38 (retrospective) Cases appeared in this tab when they completed the tiering pipeline Until last month, no CIP dispatch possible Case released in tiering browser to allow access for the NHS GMC Case closed in portal -> won’t be dispatched to a CIP Once CIP live on build 38, cases which haven’t been closed are being dispatched Moves cases back to tab 1 Once released in CIP system, moves back to tab 2 Considered formally released to the NHS GMC Pathway adapted to cope with a temporary situation where build 38 genomes couldn’t be processed via a CIP November 2020 39

Notes on build 38 backlog cases • When a case is re-released in the CIP system, the case ID will change, e. g. from GEL-123 to SAP-123 or OPA-123 • If NHS GMCs have cases they are working on which they don’t want sent to the CIP, please put case IDs in a service desk ticket (pending portal developments to allow this to happen directly) November 2020 40

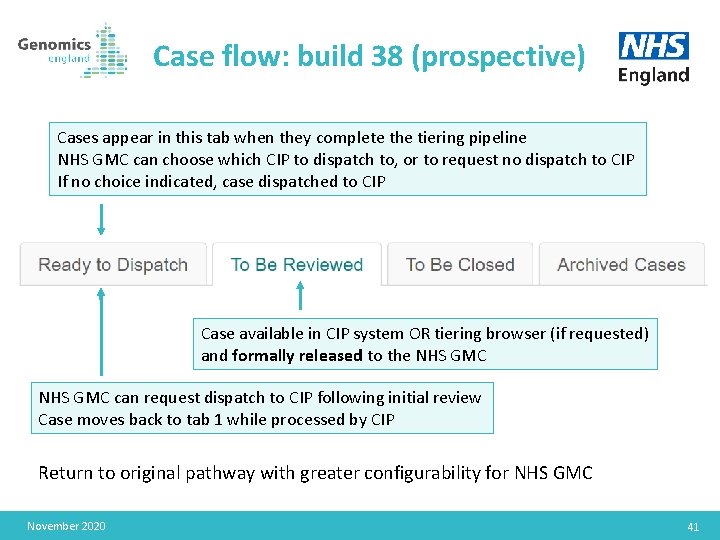
Case flow: build 38 (prospective) Cases appear in this tab when they complete the tiering pipeline NHS GMC can choose which CIP to dispatch to, or to request no dispatch to CIP If no choice indicated, case dispatched to CIP Case available in CIP system OR tiering browser (if requested) and formally released to the NHS GMC can request dispatch to CIP following initial review Case moves back to tab 1 while processed by CIP Return to original pathway with greater configurability for NHS GMC November 2020 41

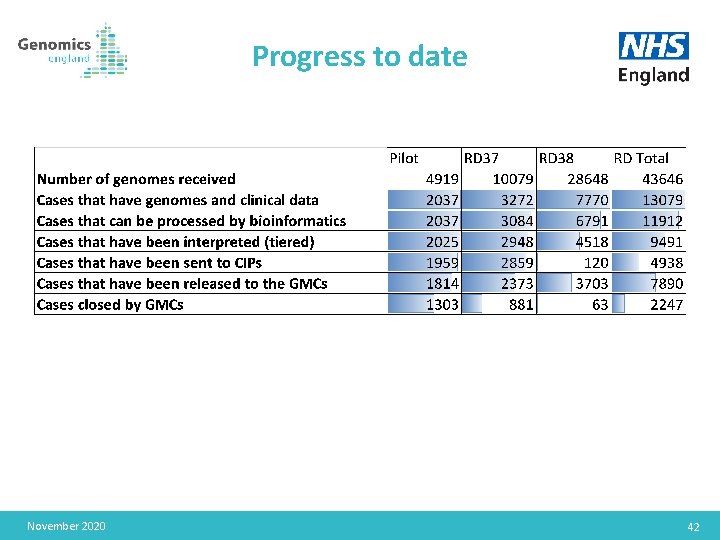
Progress to date November 2020 42

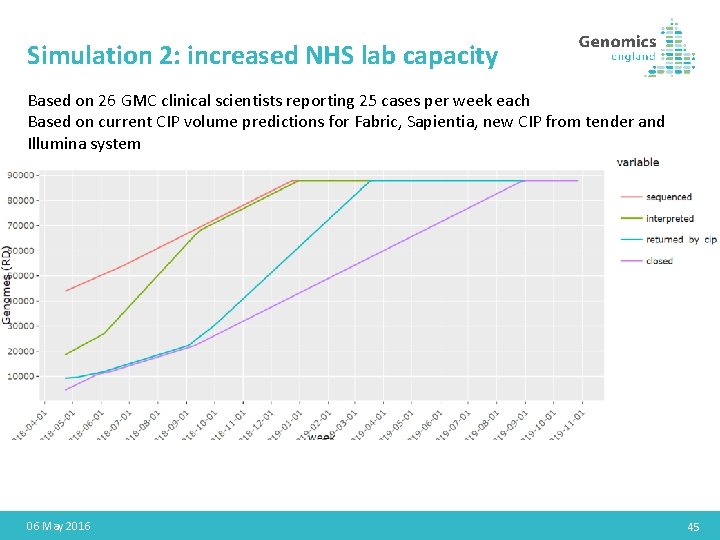
Results processing modelling • Models have been created for build 38 rare disease results • Assumptions can be challenged and other scenarios modelled easily • Models only currently consider 100 k results; need updating to consider interplay between 100 k results and early GMS results processing • NHSE keen to use modelling to work with NHS GMCs for workforce planning and resource allocation • National approaches to staff training and support for labs could be considered if helpful November 2020 43

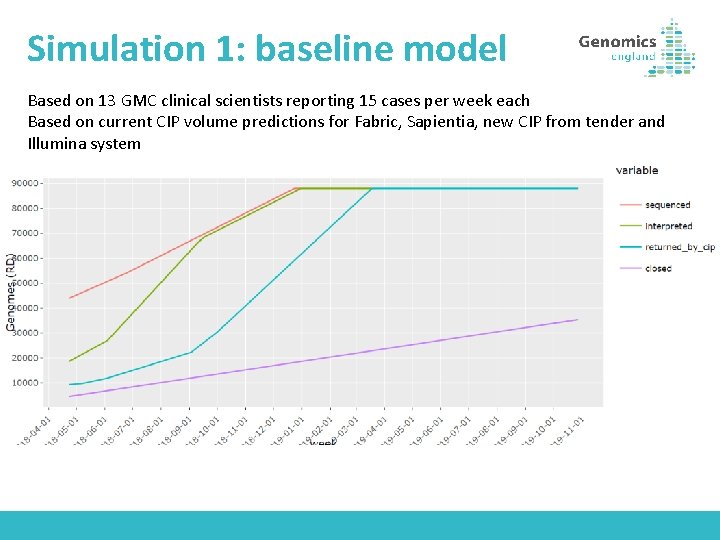
Simulation 1: baseline model Based on 13 GMC clinical scientists reporting 15 cases per week each Based on current CIP volume predictions for Fabric, Sapientia, new CIP from tender and Illumina system

Simulation 2: increased NHS lab capacity Based on 26 GMC clinical scientists reporting 25 cases per week each Based on current CIP volume predictions for Fabric, Sapientia, new CIP from tender and Illumina system 06 May 2016 45

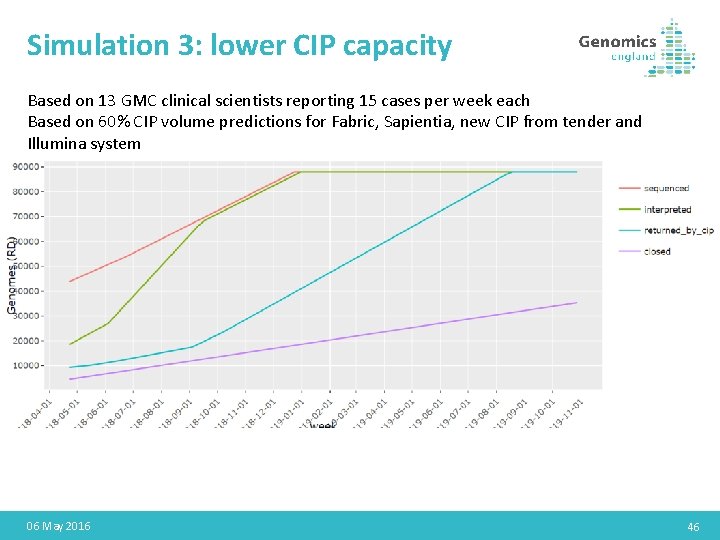
Simulation 3: lower CIP capacity Based on 13 GMC clinical scientists reporting 15 cases per week each Based on 60% CIP volume predictions for Fabric, Sapientia, new CIP from tender and Illumina system 06 May 2016 46

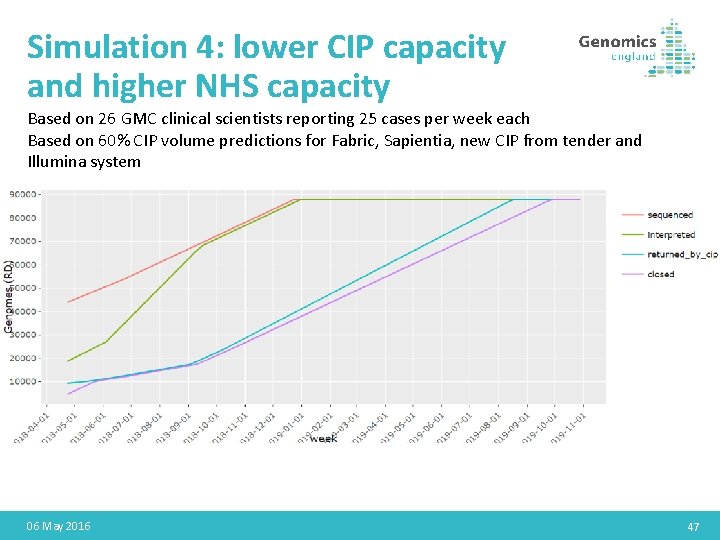
Simulation 4: lower CIP capacity and higher NHS capacity Based on 26 GMC clinical scientists reporting 25 cases per week each Based on 60% CIP volume predictions for Fabric, Sapientia, new CIP from tender and Illumina system 06 May 2016 47

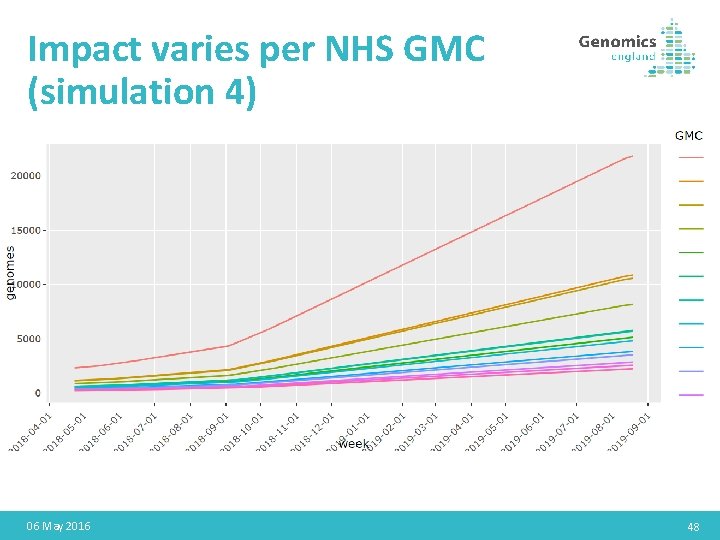
Impact varies per NHS GMC (simulation 4) 06 May 2016 48

New pipeline elements initially introduced prospectively 06 May 2016 49

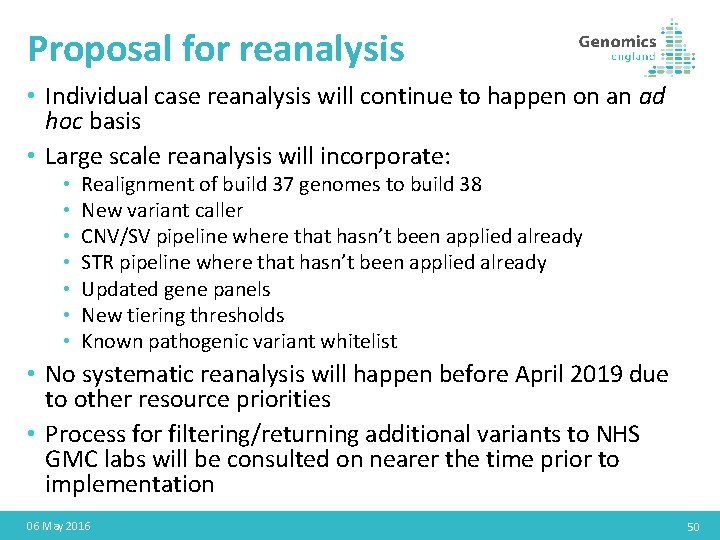
Proposal for reanalysis • Individual case reanalysis will continue to happen on an ad hoc basis • Large scale reanalysis will incorporate: • • Realignment of build 37 genomes to build 38 New variant caller CNV/SV pipeline where that hasn’t been applied already STR pipeline where that hasn’t been applied already Updated gene panels New tiering thresholds Known pathogenic variant whitelist • No systematic reanalysis will happen before April 2019 due to other resource priorities • Process for filtering/returning additional variants to NHS GMC labs will be consulted on nearer the time prior to implementation 06 May 2016 50

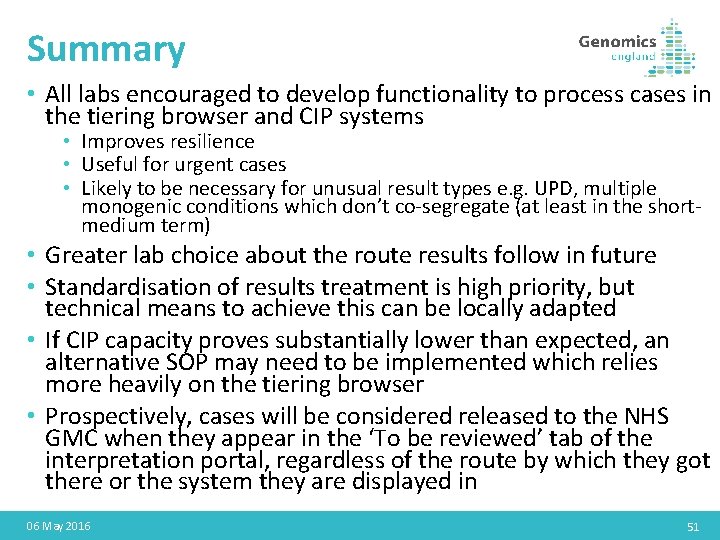
Summary • All labs encouraged to develop functionality to process cases in the tiering browser and CIP systems • Improves resilience • Useful for urgent cases • Likely to be necessary for unusual result types e. g. UPD, multiple monogenic conditions which don’t co-segregate (at least in the shortmedium term) • Greater lab choice about the route results follow in future • Standardisation of results treatment is high priority, but technical means to achieve this can be locally adapted • If CIP capacity proves substantially lower than expected, an alternative SOP may need to be implemented which relies more heavily on the tiering browser • Prospectively, cases will be considered released to the NHS GMC when they appear in the ‘To be reviewed’ tab of the interpretation portal, regardless of the route by which they got there or the system they are displayed in 06 May 2016 51

Validation update 52

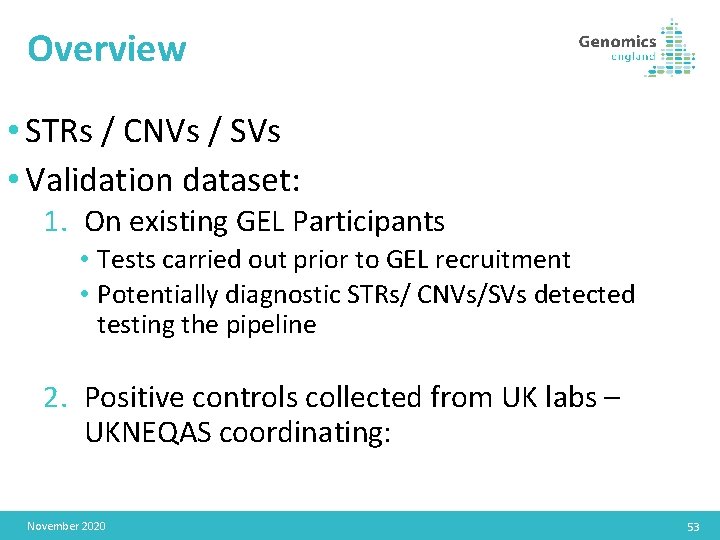
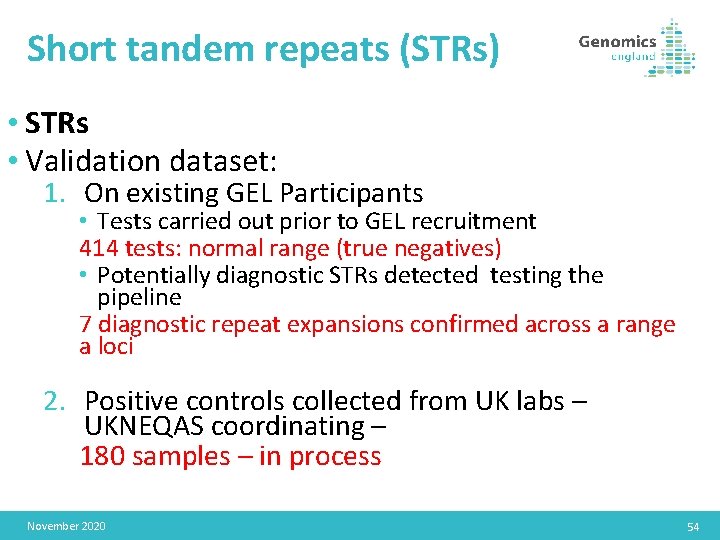
Overview • STRs / CNVs / SVs • Validation dataset: 1. On existing GEL Participants • Tests carried out prior to GEL recruitment • Potentially diagnostic STRs/ CNVs/SVs detected testing the pipeline 2. Positive controls collected from UK labs – UKNEQAS coordinating: November 2020 53

Short tandem repeats (STRs) • STRs • Validation dataset: 1. On existing GEL Participants • Tests carried out prior to GEL recruitment 414 tests: normal range (true negatives) • Potentially diagnostic STRs detected testing the pipeline 7 diagnostic repeat expansions confirmed across a range a loci 2. Positive controls collected from UK labs – UKNEQAS coordinating – 180 samples – in process November 2020 54

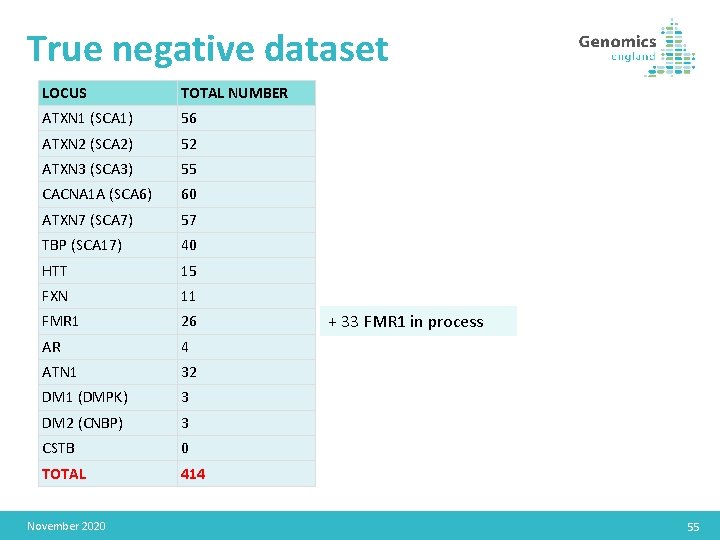
True negative dataset LOCUS TOTAL NUMBER ATXN 1 (SCA 1) 56 ATXN 2 (SCA 2) 52 ATXN 3 (SCA 3) 55 CACNA 1 A (SCA 6) 60 ATXN 7 (SCA 7) 57 TBP (SCA 17) 40 HTT 15 FXN 11 FMR 1 26 AR 4 ATN 1 32 DM 1 (DMPK) 3 DM 2 (CNBP) 3 CSTB 0 TOTAL 414 November 2020 + 33 FMR 1 in process 55

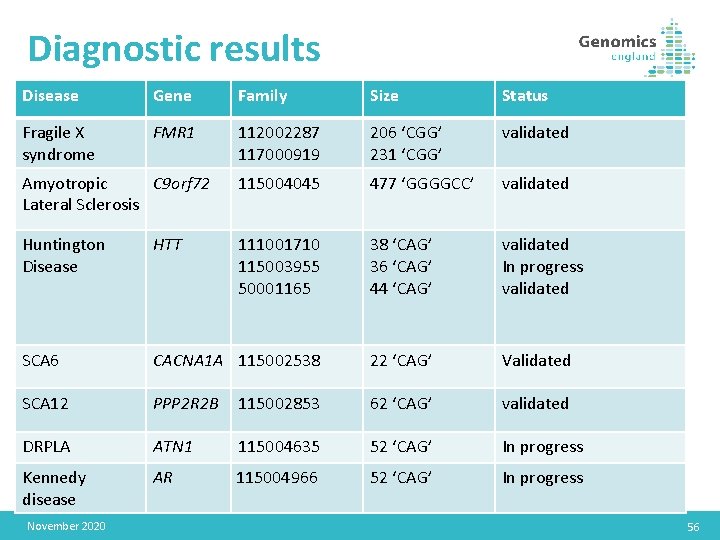
Diagnostic results Disease Gene Family Size Status Fragile X syndrome FMR 1 112002287 117000919 206 ‘CGG’ 231 ‘CGG’ validated Amyotropic C 9 orf 72 Lateral Sclerosis 115004045 477 ‘GGGGCC’ validated Huntington Disease HTT 111001710 115003955 50001165 38 ‘CAG’ 36 ‘CAG’ 44 ‘CAG’ validated In progress validated SCA 6 CACNA 1 A 115002538 22 ‘CAG’ Validated SCA 12 PPP 2 R 2 B 115002853 62 ‘CAG’ validated DRPLA ATN 1 115004635 52 ‘CAG’ In progress Kennedy disease AR 115004966 52 ‘CAG’ In progress November 2020 56

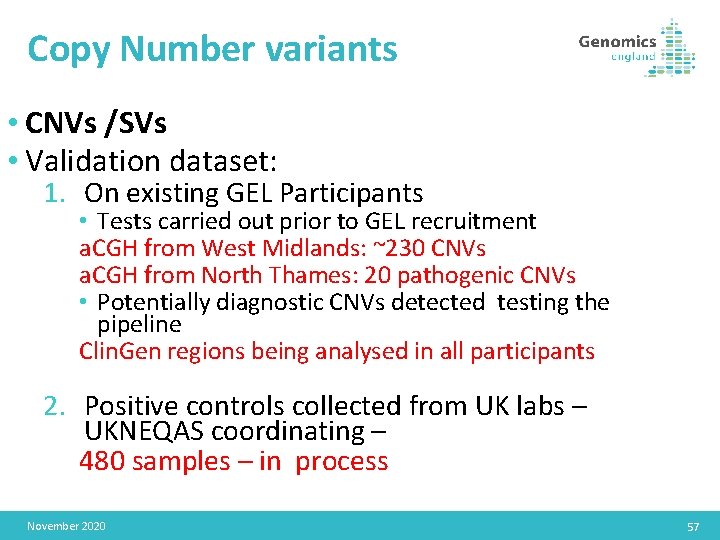
Copy Number variants • CNVs /SVs • Validation dataset: 1. On existing GEL Participants • Tests carried out prior to GEL recruitment a. CGH from West Midlands: ~230 CNVs a. CGH from North Thames: 20 pathogenic CNVs • Potentially diagnostic CNVs detected testing the pipeline Clin. Gen regions being analysed in all participants 2. Positive controls collected from UK labs – UKNEQAS coordinating – 480 samples – in process November 2020 57

Small variants in ‘tricky’ genes • PKD 1 • validation dataset: 1. On existing GEL Participants • Potentially diagnostic PKD 1 variants detected testing the pipeline 22 families in GEL that had the PKD panel applied: • 13 of which have either tier 1 or tier 2 variants • 9 have no tiered variants. • Cambridge lab will confirm T 1/T 2 variants 2. Positive controls collected from UK labs – UKNEQAS coordinating – Cambridge lab (Ed Thompson) and Other labs will contribute positive controls November 2020 58