Genomics Core Lab Department of Pathology Brody School

Genomics Core Lab Department of Pathology Brody School of Medicine East Carolina University
Genomics Education Our Goals: Research Service To serve the ECU community with the cutting-edge NGS technologies, genomics and bioinformatics.

Our Sequencers: • Mi. Seq (Illumina) 15 - 25 M short-reads per sequencing run

Our Sequencers: • Mi. Seq (Illumina) • Next. Seq 2000 (Illumina) 400 - 1000 M short-reads per sequencing run

Our Sequencers: • Mi. Seq (Illumina) • Next. Seq 2000 (Illumina) • Sequel IIe (Pacific Biosciences)

Our Liquid Handling Automations: • Biomek i 7 (Beckman Coulter) high-throughput with 96 channel head and span-8

Our Liquid Handling Automations: • Biomek i 7 (Beckman Coulter) • ep. Motion 5075 (Eppendorf) flexible & integrated

Our Liquid Handling Automations: • Biomek i 7 (Beckmen Coulter) • ep. Motion 5075 (Eppendorf) • Mantis (Formulatrix) microfluidic liquid dispenser
Our Single-Cell Analysis: • gentle. MACS Octo Dissociator with heaters (Miltenyi Biotec) semi-automated and standardized tissue dissociation and single-cell separation
Our Single-Cell Analysis: • gentle. MACS Octo Dissociator with heaters (Miltenyi Biotec) • Chromium Controller (10 x Genomics) single-cell RNA sequencing (sc. RNAseq) single-cell epigenomic profiling (ATAC-seq) single-cell immune profiling – V(D)J sequencing
Whole Genome Sequencing (WGS) Whole Exome Sequencing (WES) Whole Transcriptome Sequencing (RNAseq) Core Services including Bioinformatics: Gene Panel Target Sequencing Small RNA (micro. RNA) Sequencing Metagenomic Sequencing (16 S or shotgun, DNA or RNA) Single-Cell RNA Sequencing (sc. RNAseq) Single-Cell Multi-omics Sequencing (ATAC-seq & sc. RNAseq)
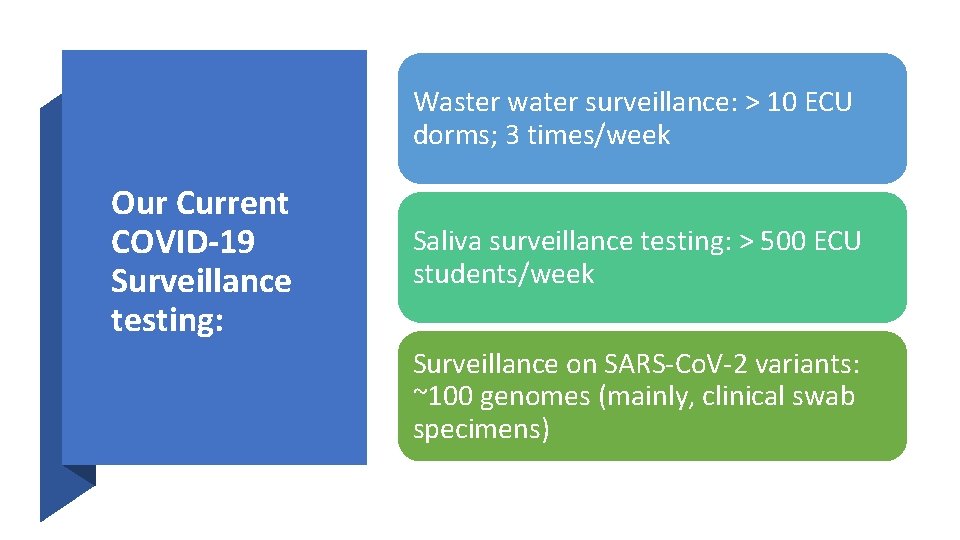
Waster water surveillance: > 10 ECU dorms; 3 times/week Our Current COVID-19 Surveillance testing: Saliva surveillance testing: > 500 ECU students/week Surveillance on SARS-Co. V-2 variants: ~100 genomes (mainly, clinical swab specimens)
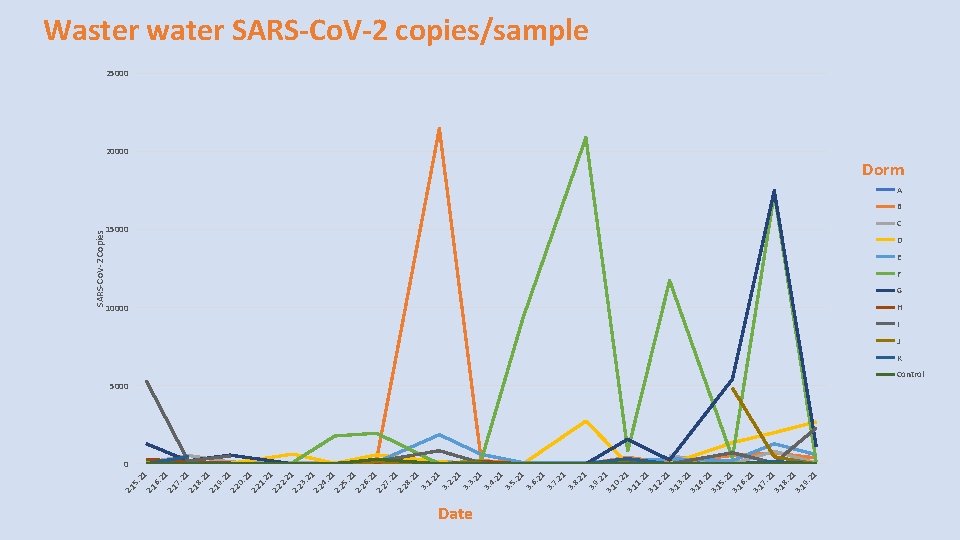
. 2 1 2. 19. 2 1 2. 20. 2 1 2. 21. 2 1 2. 23. 2 1 2. 24. 2 1 2. 25. 2 1 2. 26. 2 1 2. 27. 2 1 2. 28. 2 1 3. 1. 21 3. 2. 21 3. 3. 21 3. 4. 21 3. 5. 21 3. 6. 21 3. 7. 21 3. 8. 21 3. 9. 21 3. 10. 2 1 3. 11. 2 1 3. 12. 2 1 3. 13. 2 1 3. 14. 2 1 3. 15. 2 1 3. 16. 2 1 3. 17. 2 1 3. 18. 2 1 3. 19. 2 1 2. 18 2. 17 2. 16 2. 15 SARS-Co. V-2 Copies Waster water SARS-Co. V-2 copies/sample 25000 20000 Dorm A B 15000 C D E F G 10000 H I J K 5000 Control 0 Date
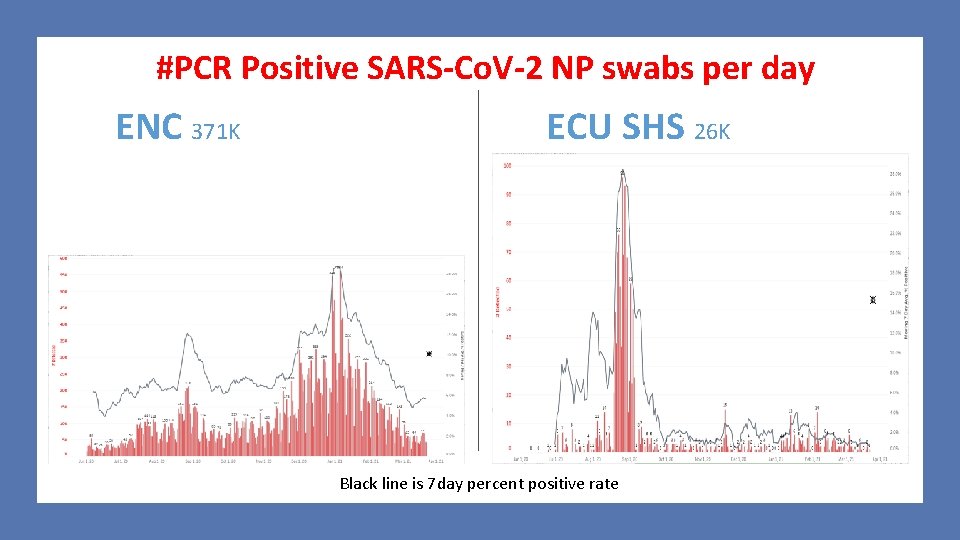
#PCR Positive SARS-Co. V-2 NP swabs per day ENC 371 K ECU SHS 26 K Black line is 7 day percent positive rate
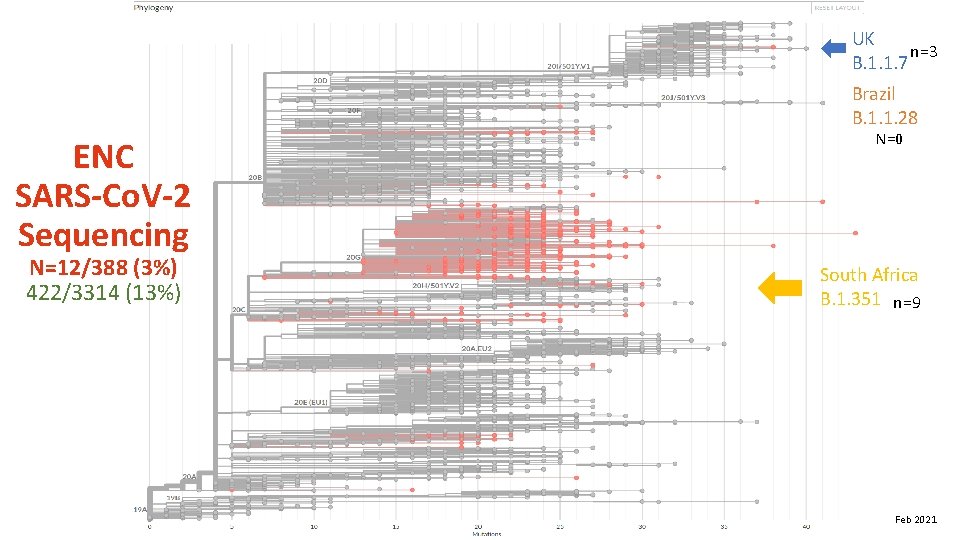
UK n=3 B. 1. 1. 7 Brazil B. 1. 1. 28 ENC SARS-Co. V-2 Sequencing N=12/388 (3%) 422/3314 (13%) N=0 South Africa B. 1. 351 n=9 Feb 2021
We will provide the best Genomic service possible to all of ECU. Due to high complexity of NGS technologies, methodologies, and bioinformatics analyses, consultation is highly recommended before starting your projects. For methodology incorporation and protocol optimization to meet your research requirements and demands, collaboration is highly encouraged.

Genomics Core Lab Department of Pathology Brody 7 S Brody School of Medicine East Carolina University Contact Info: John T. Fallon, MD, Ph. D Weihua Huang, Ph. D Changhong Yin, MD, Ph. D Kim Briley, MS fallonj 19@ecu. edu huangw 20@ecu. edu yinc 20@ecu. edu brileyki@ecu. edu
- Slides: 18