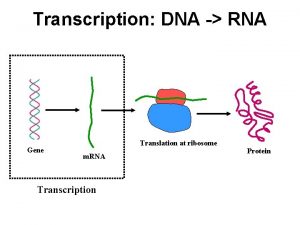
Genomewide analysis of enhancerRNA transcription reveals regulatory mechanisms







- Slides: 28

Genome-wide analysis of enhancer-RNA transcription reveals regulatory mechanisms by an anti-diabetic drugs (TZD) in adipocytes Kyoung-Jae Won, Ph. D. Sep 8 2014 Genomics 2014

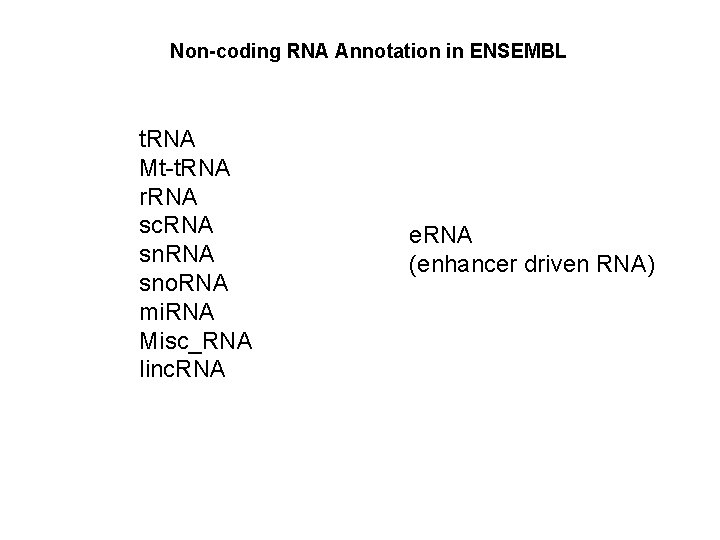
Non-coding RNA Annotation in ENSEMBL t. RNA Mt-t. RNA r. RNA sc. RNA sno. RNA mi. RNA Misc_RNA linc. RNA e. RNA (enhancer driven RNA)

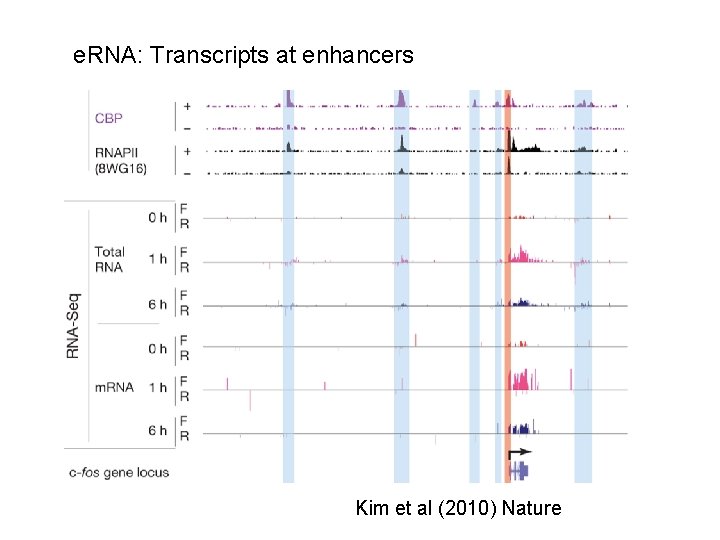
e. RNA: Transcripts at enhancers Discussions Kim et al (2010) Nature

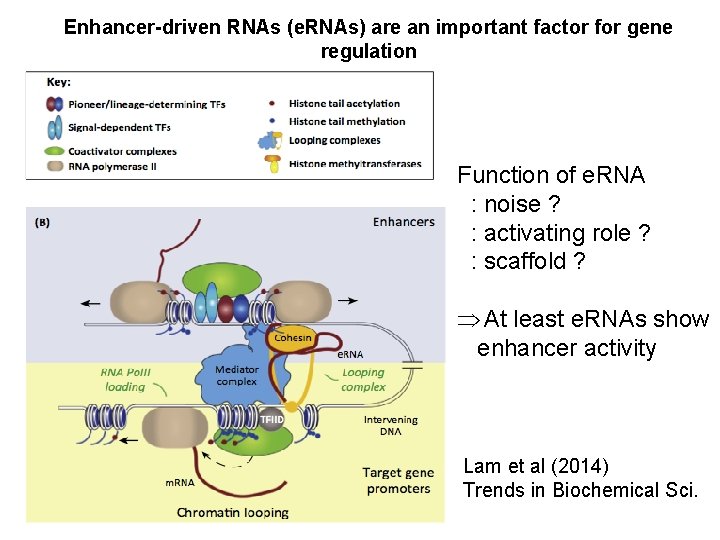
Enhancer-driven RNAs (e. RNAs) are an important factor for gene regulation Function of e. RNA : noise ? : activating role ? : scaffold ? At least e. RNAs show enhancer activity Lam et al (2014) Trends in Biochemical Sci.

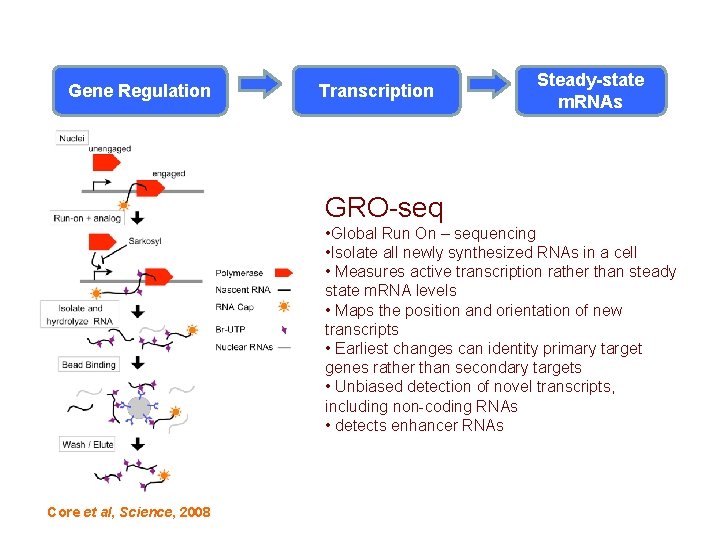
Transcription: An Understudied Step in Gene Regulation Transcription Steady-state m. RNAs GRO-seq • Global Run On – sequencing • Isolate all newly synthesized RNAs in a cell • Measures active transcription rather than steady state m. RNA levels • Maps the position and orientation of new transcripts • Earliest changes can identity primary target genes rather than secondary targets • Unbiased detection of novel transcripts, including non-coding RNAs • detects enhancer RNAs Core et al, Science, 2008

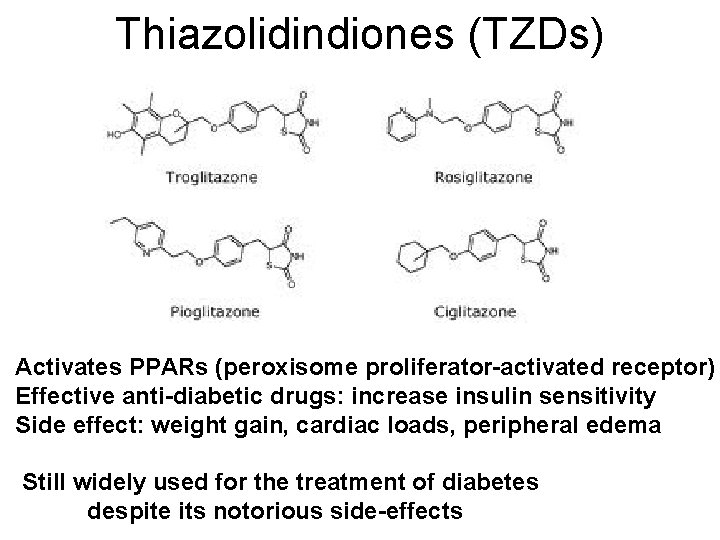
Thiazolidindiones (TZDs) Activates PPARs (peroxisome proliferator-activated receptor) Effective anti-diabetic drugs: increase insulin sensitivity Side effect: weight gain, cardiac loads, peripheral edema Still widely used for the treatment of diabetes despite its notorious side-effects

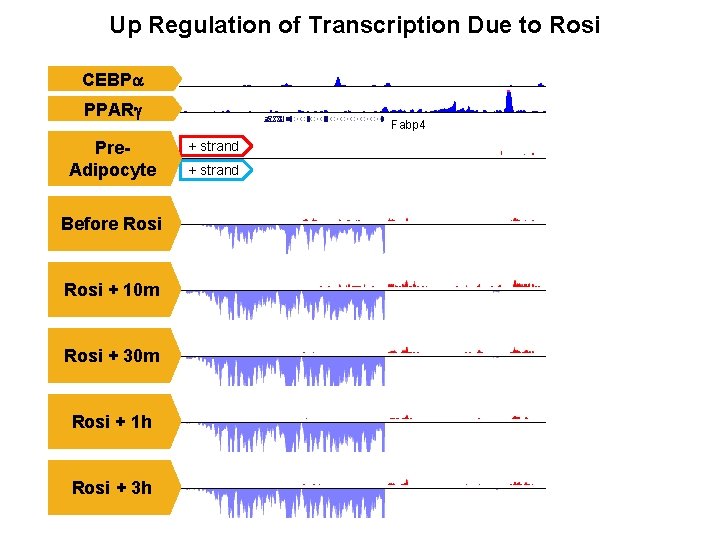
Up Regulation of Transcription Due to Rosi CEBPa PPARg Pre. Adipocyte Before Rosi + 10 m Rosi + 30 m Rosi + 1 h Rosi + 3 h Fabp 4 + strand

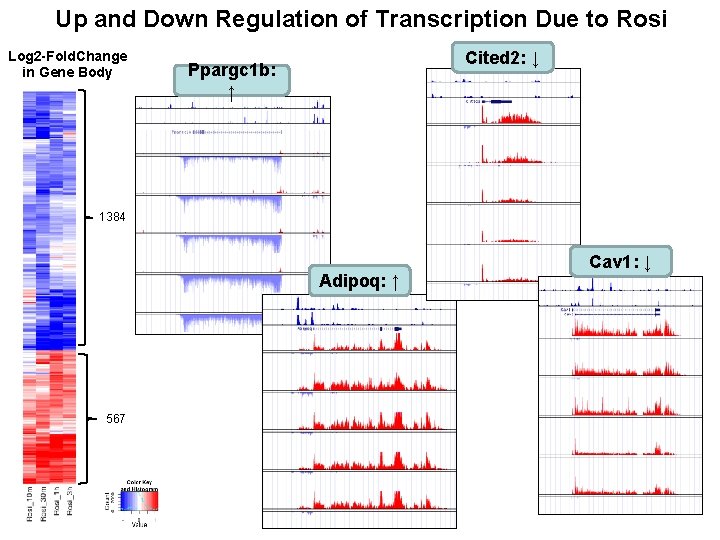
Up and Down Regulation of Transcription Due to Rosi Log 2 -Fold. Change in Gene Body Cited 2: ↓ Ppargc 1 b: ↑ 1384 Adipoq: ↑ 567 Cav 1: ↓

GROseq Profiles at TSS in Adipocyte

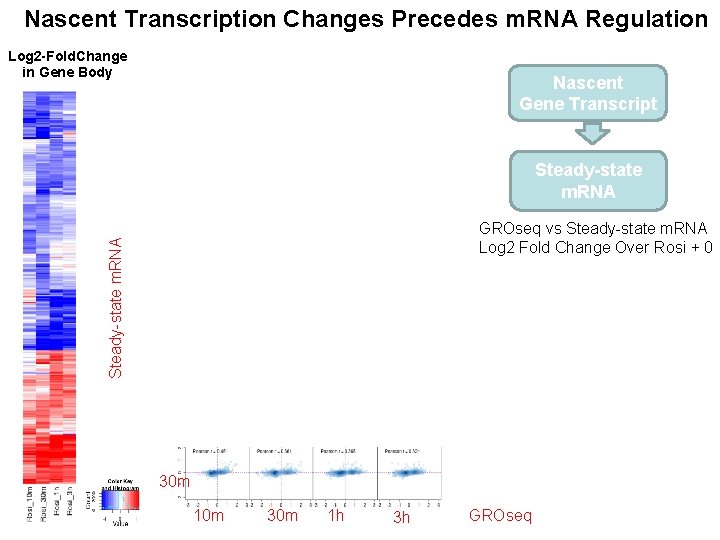
Nascent Transcription Changes Precedes m. RNA Regulation Log 2 -Fold. Change in Gene Body Nascent Gene Transcript Steady-state m. RNA GROseq vs Steady-state m. RNA Log 2 Fold Change Over Rosi + 0 30 m 10 m 30 m 1 h 3 h GROseq

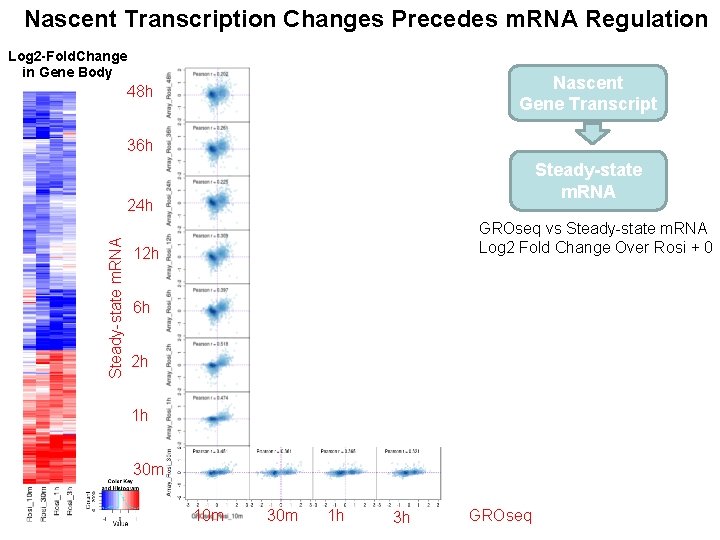
Nascent Transcription Changes Precedes m. RNA Regulation Log 2 -Fold. Change in Gene Body Nascent Gene Transcript 48 h 36 h Steady-state m. RNA 24 h GROseq vs Steady-state m. RNA Log 2 Fold Change Over Rosi + 0 12 h 6 h 2 h 1 h 30 m 10 m 30 m 1 h 3 h GROseq

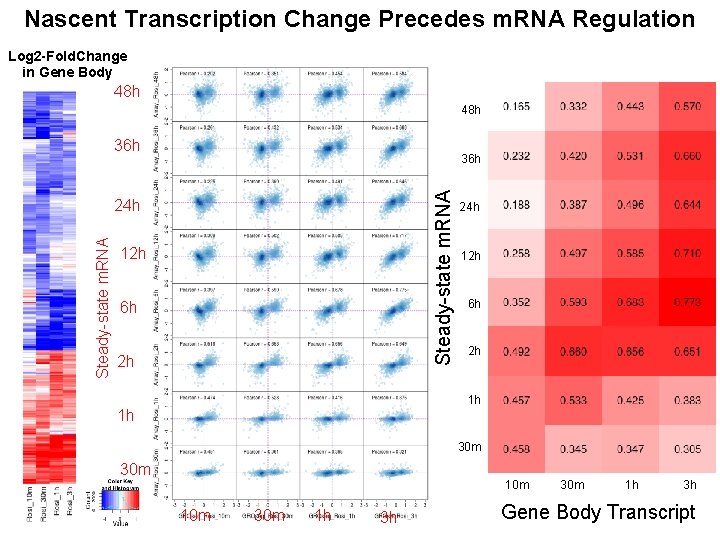
Nascent Transcription Change Precedes m. RNA Regulation Log 2 -Fold. Change in Gene Body 48 h 36 h Steady-state m. RNA 24 h 12 h 6 h 2 h 1 h 1 h 30 m 10 m 30 m 1 h 3 h Gene Body Transcript

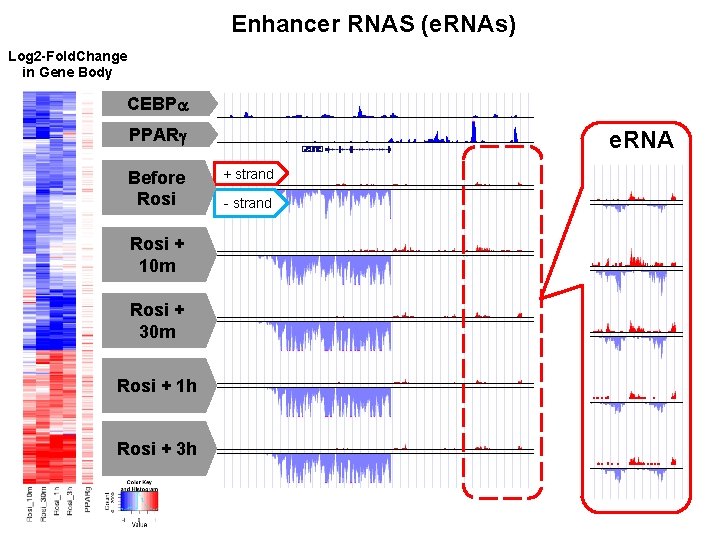
Enhancer RNAS (e. RNAs) Log 2 -Fold. Change in Gene Body CEBPa PPARg Before Rosi + 10 m Rosi + 30 m Rosi + 1 h Rosi + 3 h e. RNA + strand - strand

Rosi-regulated e. RNAs

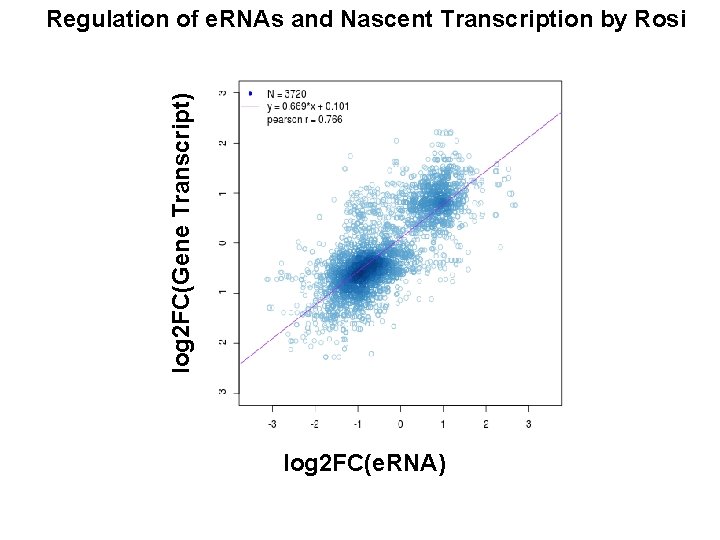
log 2 FC(Gene Transcript) Regulation of e. RNAs and Nascent Transcription by Rosi log 2 FC(e. RNA)

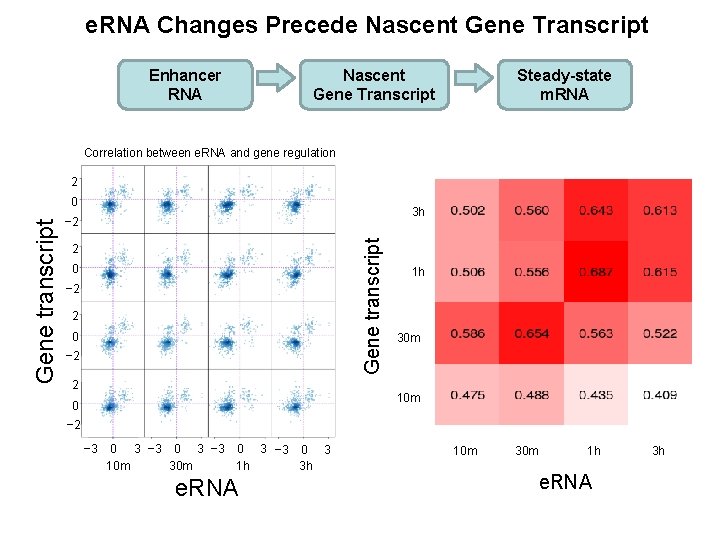
e. RNA Changes Precede Nascent Gene Transcript Enhancer RNA Nascent Gene Transcript Steady-state m. RNA Correlation between e. RNA and gene regulation 2 3 h − 2 Gene transcript 0 2 0 − 2 2 1 h 30 m 10 m 0 − 2 − 3 0 3 10 m 30 m 1 h 3 h e. RNA 10 m 30 m 1 h e. RNA 3 h

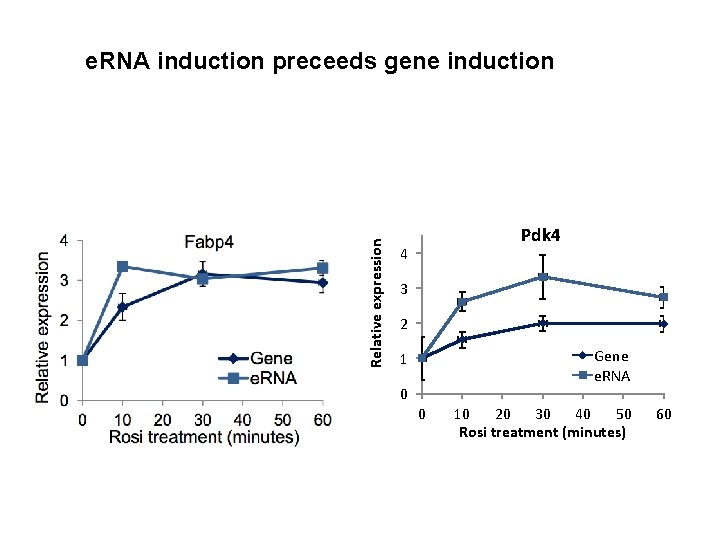
Relative expression e. RNA induction preceeds gene induction Pdk 4 4 3 2 Gene e. RNA 1 0 0 10 20 30 40 50 Rosi treatment (minutes) 60

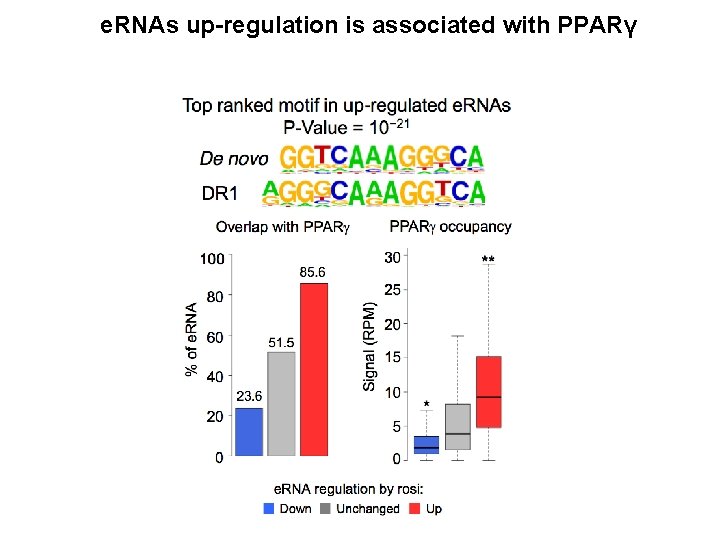
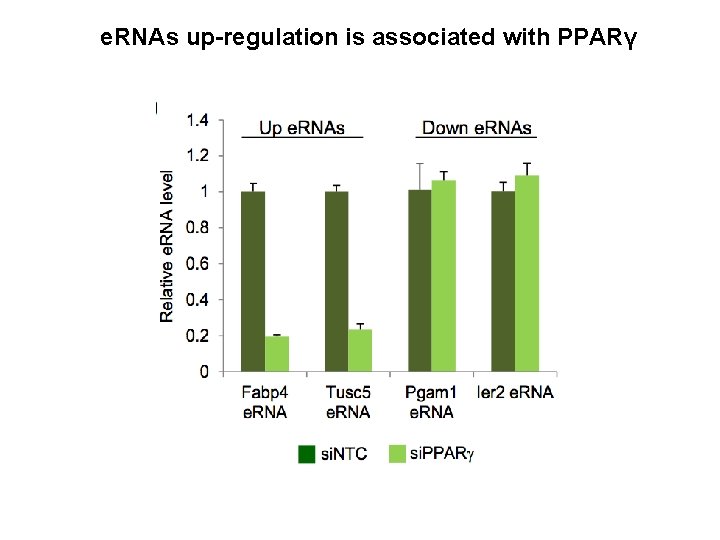
e. RNAs up-regulation is associated with PPARγ

e. RNAs up-regulation is associated with PPARγ

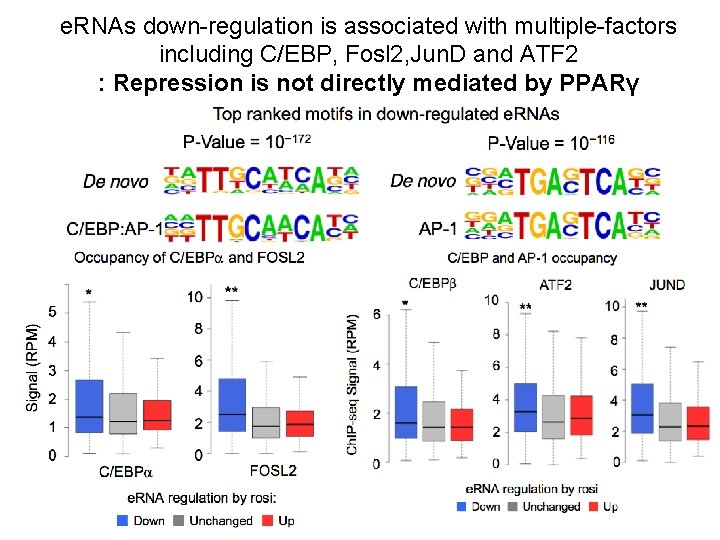
e. RNAs down-regulation is associated with multiple-factors including C/EBP, Fosl 2, Jun. D and ATF 2 : Repression is not directly mediated by PPARγ

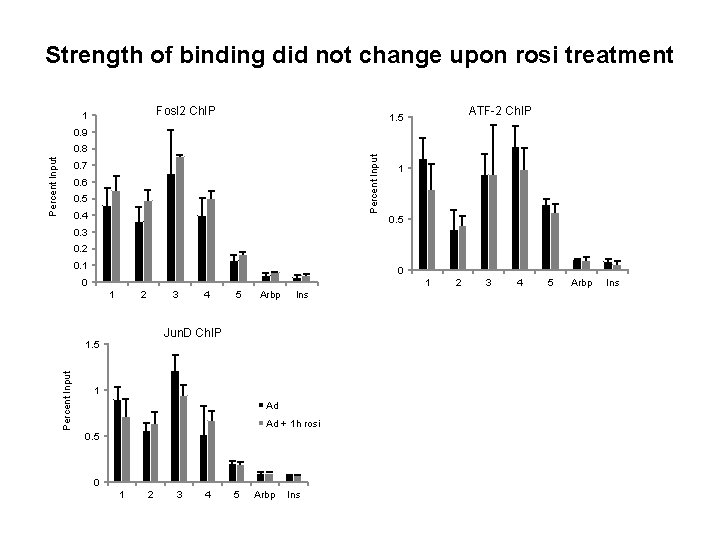
Strength of binding did not change upon rosi treatment Fosl 2 Ch. IP 1 ATF-2 Ch. IP 1. 5 0. 9 Percent Input 0. 8 0. 7 0. 6 0. 5 0. 4 1 0. 5 0. 3 0. 2 0. 1 0 0 1 1 2 3 5 Arbp Ins Jun. D Ch. IP 1. 5 Percent Input 4 1 Ad Ad + 1 h rosi 0. 5 0 1 2 3 4 5 Arbp Ins

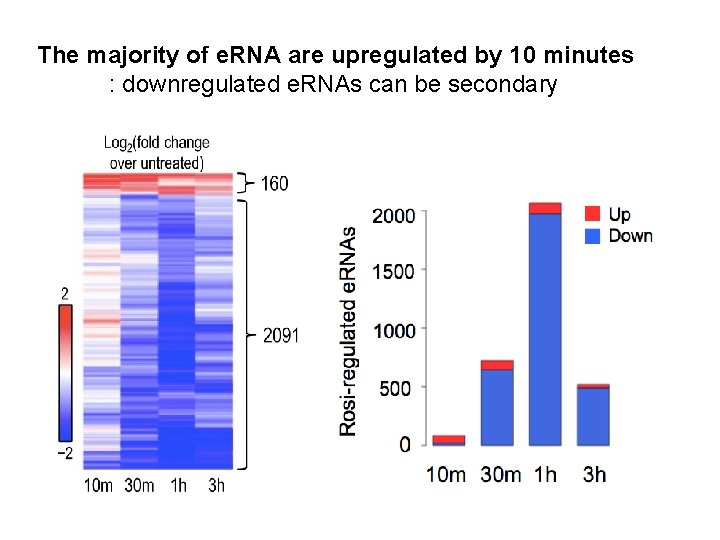
The majority of e. RNA are upregulated by 10 minutes : downregulated e. RNAs can be secondary

e. RNA changes are associated with Med 1 change

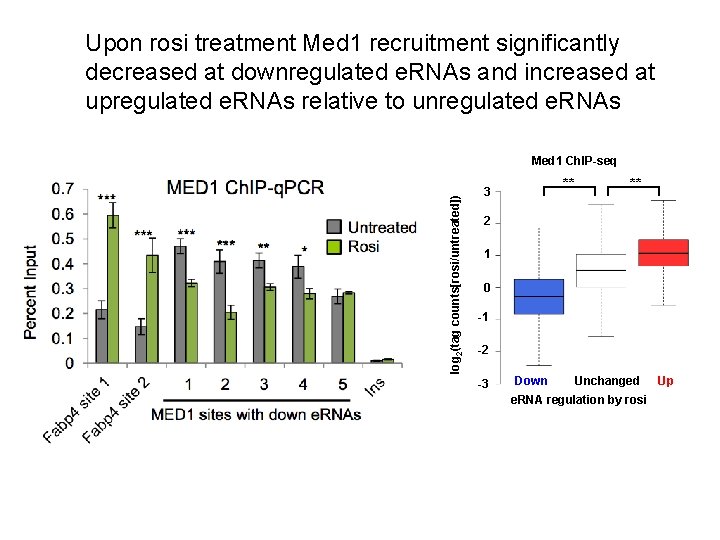
Upon rosi treatment Med 1 recruitment significantly decreased at downregulated e. RNAs and increased at upregulated e. RNAs relative to unregulated e. RNAs log 2(tag counts[rosi/untreated]) Med 1 Ch. IP-seq ** 3 ** 2 1 0 -1 -2 -3 Down Unchanged e. RNA regulation by rosi Up

Squelching is a biological phenomenon in which a transcriptional activator acts to inhibit the expression of another gene.

Discussions GROseq measures the level of e. RNAs and gene transcript Integrative studies using GROseq revealed the rosi-associated regulatory mechanisms • Gene transcription levels needs time until they reach to steady state levels • e. RNA fires before transcriptional initiation • up-regulation is mediated by PPARγ • Multiple TFs are associated with down-regulation • down-regulation is mediated by co-activator redistribution Step et al. (2014) Genes and Development

Thanks to Nha Nguyen Hee-Woong Lim Shuo Li Inchan Choi UPENN Mitchell Lazar Sonia Stem Patrick Seale David Steger Klaus Kaestner Rinho Kim Supported by • NIH/NIDDK, • Institute for Diabetes Obesity and Metabolism at the University of Pennsylvania • Pharma Foundation

In the lab. . Metabolic Biology Mitchell Lazar, Klaus Keastner Patrick Seale, David Steger Stem Cell Kennett Zaret, Montserrat Aguera Chromatin remodelers, histone variants Benjamin Garcia, Hua-Ying Epigenetic landscapes Wei Wang, Bing Ren (UCSD) Cancer biology Cathryn Wellen, Mariuz Wasik Integrative studies RNAseq, GROseq Ch. IPseq, Ch. IP-exo CLIP-seq m. C, 5 hm. C, Histone modification
 Define central idea
Define central idea Art reveals society
Art reveals society Mark twain reveals stage fright
Mark twain reveals stage fright The way a writer reveals the personality of a character.
The way a writer reveals the personality of a character. And think this heart all evil shed away
And think this heart all evil shed away The theme is the central idea or truth in the story
The theme is the central idea or truth in the story A sustainability report reveals a firm's
A sustainability report reveals a firm's Whats expository writing
Whats expository writing Examples of stage direction
Examples of stage direction Reveals that his supporters will be made earls
Reveals that his supporters will be made earls Hamlet act 4 and 5 summary
Hamlet act 4 and 5 summary Characterization is the way an author
Characterization is the way an author Transcription and translation coloring
Transcription and translation coloring Rna polymerase
Rna polymerase The sounds of language
The sounds of language Where did hiv come from
Where did hiv come from Speech phonetic transcription
Speech phonetic transcription Perbedaan replikasi virus dna dan rna
Perbedaan replikasi virus dna dan rna Transcription translation replication
Transcription translation replication Prokaryotic promoter vs eukaryotic promoter
Prokaryotic promoter vs eukaryotic promoter Progressive assimilation
Progressive assimilation Odd phonetic transcription
Odd phonetic transcription Transcription error
Transcription error Transcription translation replication
Transcription translation replication Codon wheel
Codon wheel Dna to rna transcription
Dna to rna transcription Phonetic transcription
Phonetic transcription How to write 115 on a check
How to write 115 on a check Bbc bitesize protein synthesis
Bbc bitesize protein synthesis