GenomeScale CRISPRMediated Control of Gene Repression and Activation






- Slides: 20

Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation Gilbert et al. 2014 By Lily Morache & Shahab Yazdanpanah

Introduction and Significance ● Lots of mammalian transcripts, but limited understanding of their functions ● Less known about noncoding regions (enhancers) ● Solution: alter expression levels to better understand transcript function

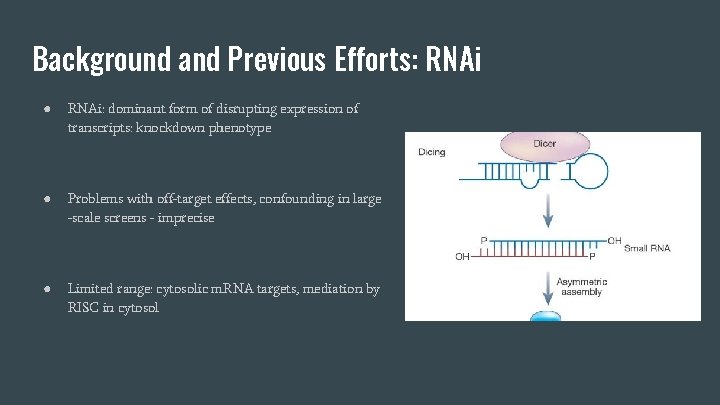
Background and Previous Efforts: RNAi ● RNAi: dominant form of disrupting expression of transcripts: knockdown phenotype ● Problems with off-target effects, confounding in large -scale screens - imprecise ● Limited range: cytosolic m. RNA targets, mediation by RISC in cytosol

● High specificity Objective ● High reproducibility ● Non-toxicity ● High efficiency ● CRISPRi/a

CRISPR Methodology - Provide synthesized sg. RNAs for target locating - Binding to Cas 9 endonuclease - Utilize as an activator or repressor via configuration or protein fusion

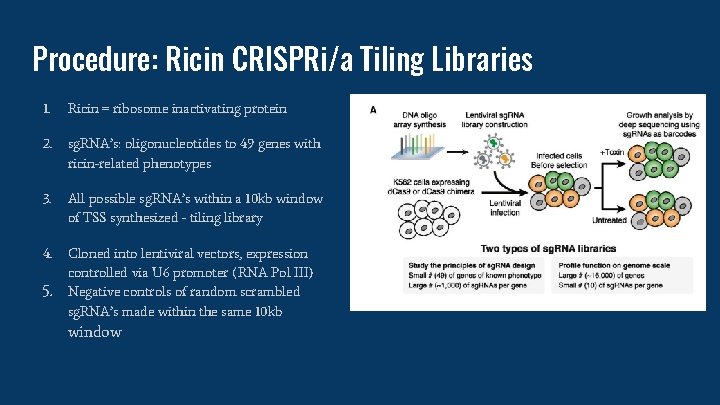
Procedure: Ricin CRISPRi/a Tiling Libraries 1. Ricin = ribosome inactivating protein 2. sg. RNA’s: oligonucleotides to 49 genes with ricin-related phenotypes 3. All possible sg. RNA’s within a 10 kb window of TSS synthesized - tiling library 4. Cloned into lentiviral vectors, expression controlled via U 6 promoter (RNA Pol III) Negative controls of random scrambled sg. RNA’s made within the same 10 kb 5. window

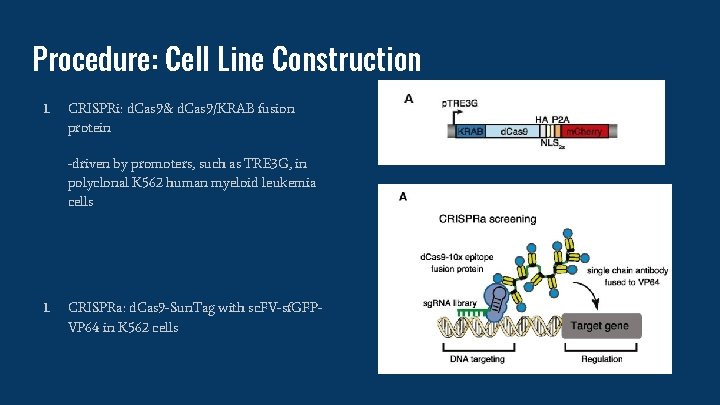
Procedure: Cell Line Construction 1. CRISPRi: d. Cas 9& d. Cas 9/KRAB fusion protein -driven by promoters, such as TRE 3 G, in polyclonal K 562 human myeloid leukemia cells 1. CRISPRa: d. Cas 9 -Sun. Tag with sc. FV-sf. GFPVP 64 in K 562 cells

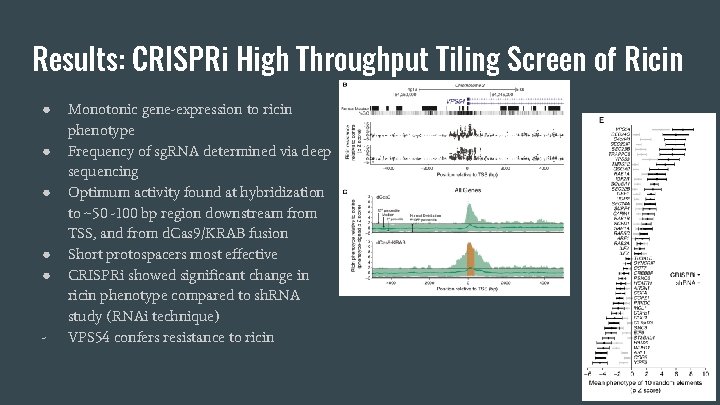
Results: CRISPRi High Throughput Tiling Screen of Ricin ● ● ● - Monotonic gene-expression to ricin phenotype Frequency of sg. RNA determined via deep sequencing Optimum activity found at hybridization to ~50 -100 bp region downstream from TSS, and from d. Cas 9/KRAB fusion Short protospacers most effective CRISPRi showed significant change in ricin phenotype compared to sh. RNA study (RNAi technique) VPS 54 confers resistance to ricin

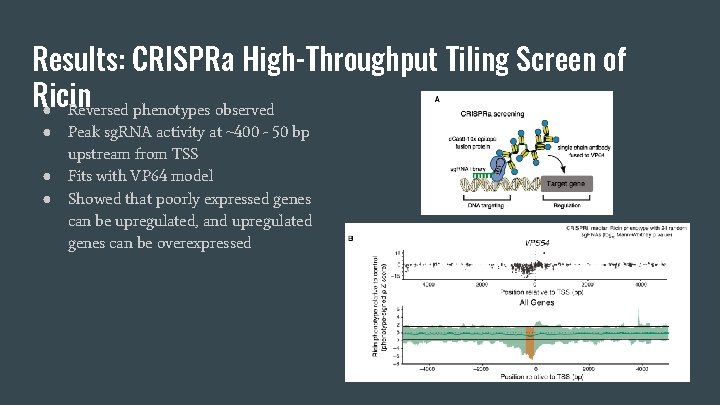
Results: CRISPRa High-Throughput Tiling Screen of Ricin ● Reversed phenotypes observed ● ● ● Peak sg. RNA activity at ~400 - 50 bp upstream from TSS Fits with VP 64 model Showed that poorly expressed genes can be upregulated, and upregulated genes can be overexpressed

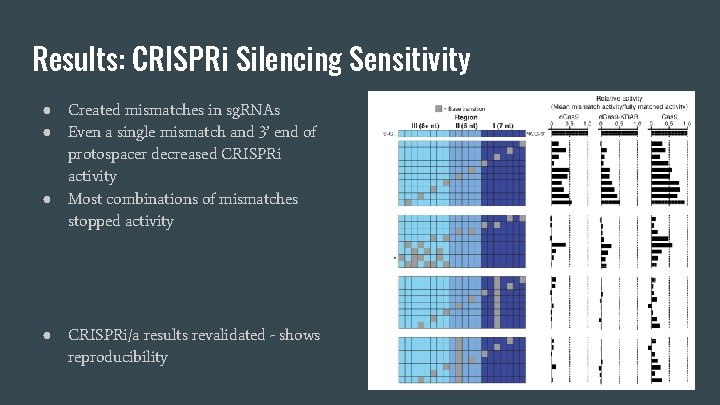
Results: CRISPRi Silencing Sensitivity ● ● Created mismatches in sg. RNAs Even a single mismatch and 3’ end of protospacer decreased CRISPRi activity Most combinations of mismatches stopped activity CRISPRi/a results revalidated - shows reproducibility

Procedure: Genome-Scale CRISPRi/a Libraries 1. Targeted 15, 977 human protein coding genes 2. 10 sg. RNA’s synthesized and designed for each gene following the tilling library, also made non -targeting control sg. RNAs 3. Total ~ 200, 000 sg. RNAs for CRISPRi/a 4. sg. RNAs and Cas 9 proteins introduced in same manner as ricin trials

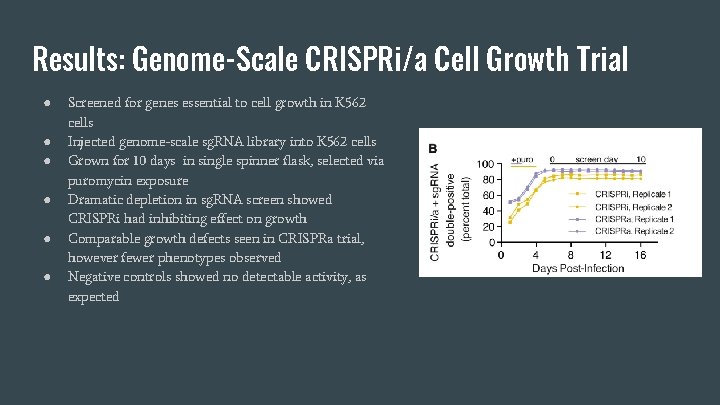
Results: Genome-Scale CRISPRi/a Cell Growth Trial ● ● ● Screened for genes essential to cell growth in K 562 cells Injected genome-scale sg. RNA library into K 562 cells Grown for 10 days in single spinner flask, selected via puromycin exposure Dramatic depletion in sg. RNA screen showed CRISPRi had inhibiting effect on growth Comparable growth defects seen in CRISPRa trial, however fewer phenotypes observed Negative controls showed no detectable activity, as expected

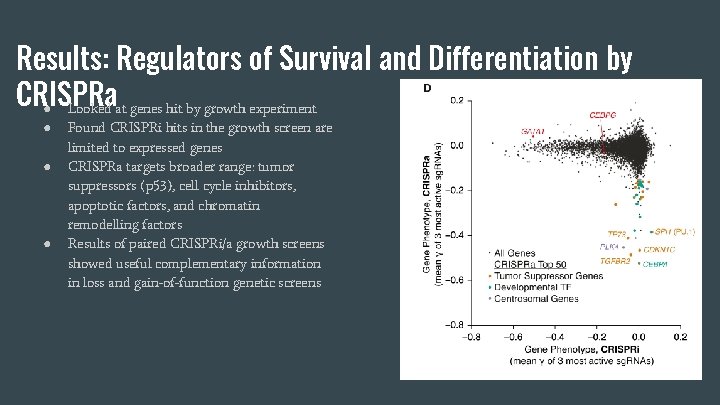
Results: Regulators of Survival and Differentiation by CRISPRa ● Looked at genes hit by growth experiment ● ● ● Found CRISPRi hits in the growth screen are limited to expressed genes CRISPRa targets broader range: tumor suppressors (p 53), cell cycle inhibitors, apoptotic factors, and chromatin remodelling factors Results of paired CRISPRi/a growth screens showed useful complementary information in loss and gain-of-function genetic screens

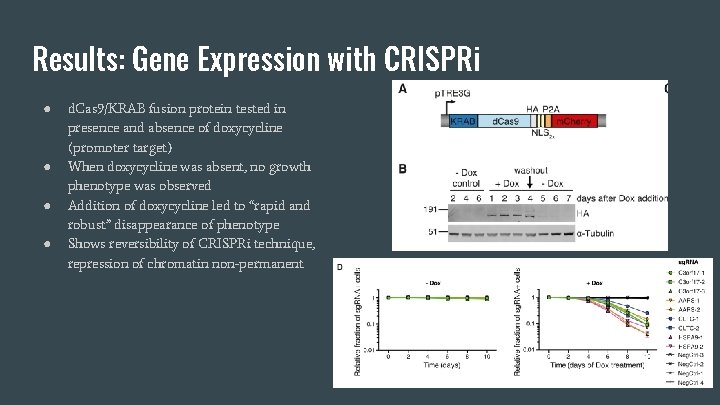
Results: Gene Expression with CRISPRi ● ● d. Cas 9/KRAB fusion protein tested in presence and absence of doxycycline (promoter target) When doxycycline was absent, no growth phenotype was observed Addition of doxycycline led to “rapid and robust” disappearance of phenotype Shows reversibility of CRISPRi technique, repression of chromatin non-permanent

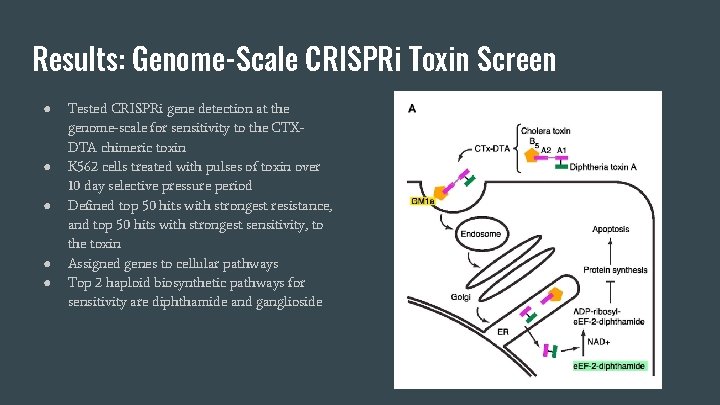
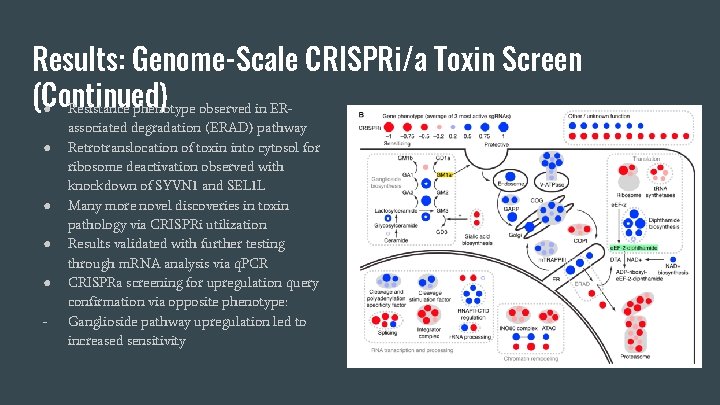
Results: Genome-Scale CRISPRi Toxin Screen ● ● ● Tested CRISPRi gene detection at the genome-scale for sensitivity to the CTXDTA chimeric toxin K 562 cells treated with pulses of toxin over 10 day selective pressure period Defined top 50 hits with strongest resistance, and top 50 hits with strongest sensitivity, to the toxin Assigned genes to cellular pathways Top 2 haploid biosynthetic pathways for sensitivity are diphthamide and ganglioside

Results: Genome-Scale CRISPRi/a Toxin Screen (Continued) ● Resistance phenotype observed in ER● ● - associated degradation (ERAD) pathway Retrotranslocation of toxin into cytosol for ribosome deactivation observed with knockdown of SYVN 1 and SEL 1 L Many more novel discoveries in toxin pathology via CRISPRi utilization Results validated with further testing through m. RNA analysis via q. PCR CRISPRa screening for upregulation query confirmation via opposite phenotype: Ganglioside pathway upregulation led to increased sensitivity

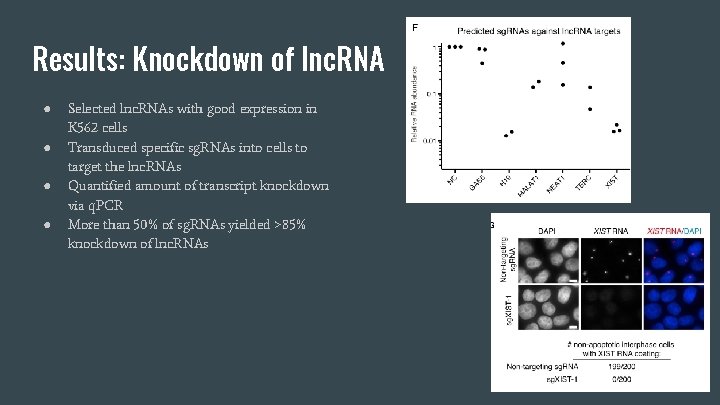
Results: Knockdown of lnc. RNA ● ● Selected lnc. RNAs with good expression in K 562 cells Transduced specific sg. RNAs into cells to target the lnc. RNAs Quantified amount of transcript knockdown via q. PCR More than 50% of sg. RNAs yielded >85% knockdown of lnc. RNAs

Discussion & Key Takeaways ● CRISPRi/a are robust tools for systematically regulating transcription of endogenous genes in human cells at the genome scale ● CRISPRi/a can be used for rapid screening of LOF and GOF phenotypes ● Identified known and unexpected genes for growth and sensitivity modulation in K 562 human myeloid leukemia cells ● CRISPRi has low instance of off-target effects, high sensitivity to mismatches ● CRISPRa can enable exploration of cellular states in which otherwise inactive pathways are induced

Improvements 1. Information was scattered throughout paper, mainly in results section 2. Redundant testing that deviated from focus of CRISPR 3. Usage of a chimeric toxin (ribosome target) may have application issues: suggest several natural toxin screens instead

Future Readings and Inquiries ● Bassik et al. , 2013; https: //www. ncbi. nlm. nih. gov/pubmed/23394947 ● Boone et al. , 2007; https: //www. ncbi. nlm. nih. gov/pubmed/17510664 ● Costanzo et al. , 2010; https: //sciencemag. org/content/327/5964/425