Genome Characterization DNA sequenceULTIMATE Map DNA sequencingmethods Assemblysequencing
Genome Characterization DNA sequence-ULTIMATE Map DNA sequencing-methods Assembly/sequencing Assigned reading: Service 2006 review paper Assigned listening: Ecic Lander genomics lecture BIO 520 Bioinformatics Jim Lund
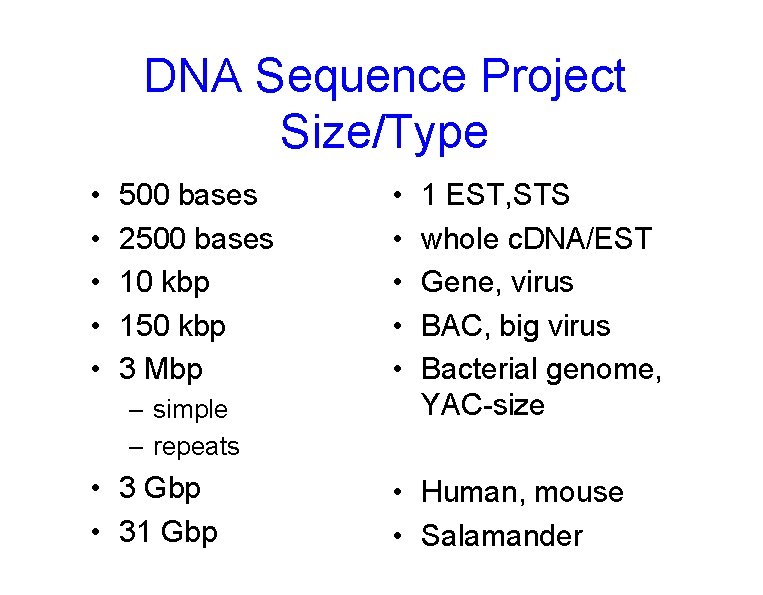
DNA Sequence Project Size/Type • • • 500 bases 2500 bases 10 kbp 150 kbp 3 Mbp – simple – repeats • 3 Gbp • 31 Gbp • • • 1 EST, STS whole c. DNA/EST Gene, virus BAC, big virus Bacterial genome, YAC-size • Human, mouse • Salamander

Metazoan genome sizes Nematode (Caenorhabditis elegans): 100 Mb Thale cress (Arabidopsis thaliana): 160 Mb Fruit fly (Drosophila melanogaster): 180 Mb Puffer fish (Takifugu rubripes): 400 Mb Rice (Oryza sativa): 490 Mb Human (Homo sapiens): 3. 5 Gb Leopard frog (Rana pipiens): 6. 5 Gb Onion (Allium cepa): 16. 4 Gb Mountain grasshopper(Podisma pedestris): 16. 5 Gb Tiger salamander (Ambystoma tigrinum): 31 Gb Easter lily (Lilium longiflorum): 34 Gb Marbled lungfish (Protopterus aethiopicus): 130 Gb

DNA Sequencing Methods • Chain termination/Dideoxy/Sanger – Fluorescence paradigm, ABI – Main method • Next generation sequencing – Polymerase addition sequencing – 454 Sequencing, Illumina – Chips: Affymetrix

Dideoxy / Chain Terminator / Sanger • Template • Primer • Extension Chemistry – polymerase – termination – labeling • Separation • Detection
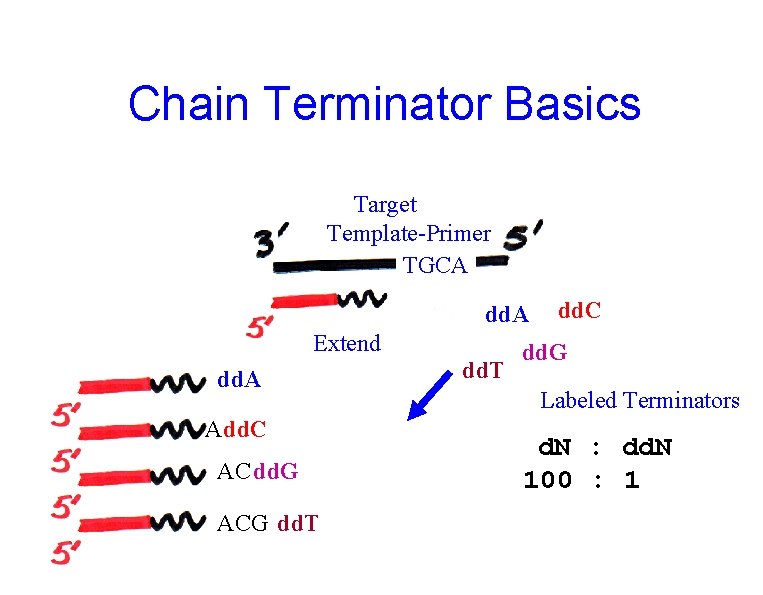
Chain Terminator Basics Target Template-Primer TGCA dd. A Extend dd. A Add. C AC dd. G ACG dd. T dd. C dd. G Labeled Terminators d. N : dd. N 100 : 1

Electrophoresis Sequencing Reaction products Polyacrylamide Gel Electrophoresis (PAGE)

DNA sequencing trace file
Separation • Gel Electrophoresis • Capillary Electrophoresis – suited to automation • • rapid (2 hrs vs 12 hrs) re-usable simple temperature control 96 well format
Paradigm Instrument • Applied Biosystems • http: //www. appliedbiosystems. com/ – ABI 3730 XL (2002, 96 samples, 1000 base reads, ~$350, 000, higher sensitivity, lower reagent cost, ~$1/reaction) – 700 Kbp / 24 hours. • 384 capillary sequencers – 5700 sequences / 24 hr day – 2. 8 Mbp / 24 hours.

384 -well capillary sequencing Results are shown as an electropherogram showing a peak for each base. From the peak heights and widths, a Phred score is assigned to each individual base. A high Phred score indicates a high certainty as to the identity of that particular base.
Sample Output 1 lane

• 1 trace=1000 bases or less – ABI: 1000 bp reads – Illumina: 50 -100 bp reads – 454 Sequencing: 300 -400 bp reads • How do we cover a genome? – DIVIDE AND CONQUER: assemble these short sequence fragments.

Assembly/Trace Editing • Consed – UNIX • EBI’s Phusion • Edit. View (ABI PRISM) – Mac • Chromas (free/pay versions) – Windows

Sequencing Strategies • Ordered – Divide and Conquer • Random Sequence – Brute Force The random approach now predominates for big projects

Random Method (details for Sanger seq) • Shear DNA (nebulize) – finish ends, ligate into vector • Produce template • Sequence to 8 X – 10 X coverage – Sequence both ends of templates. – Read length (1, 000 bp typical) – Accuracy (99% good)
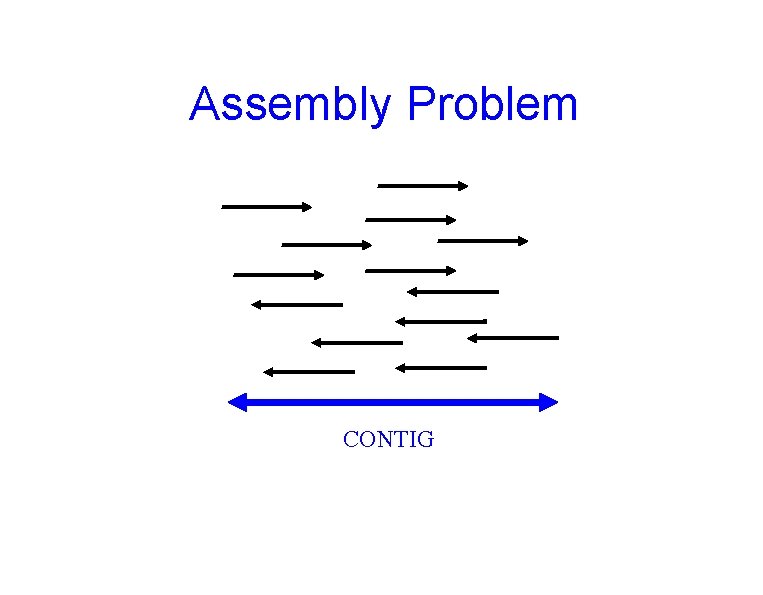
Assembly Problem CONTIG
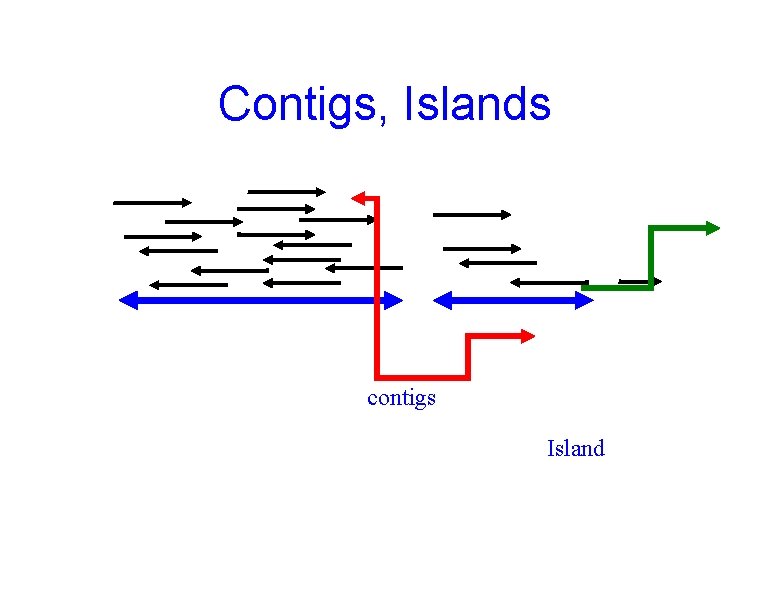
Contigs, Islands contigs Island
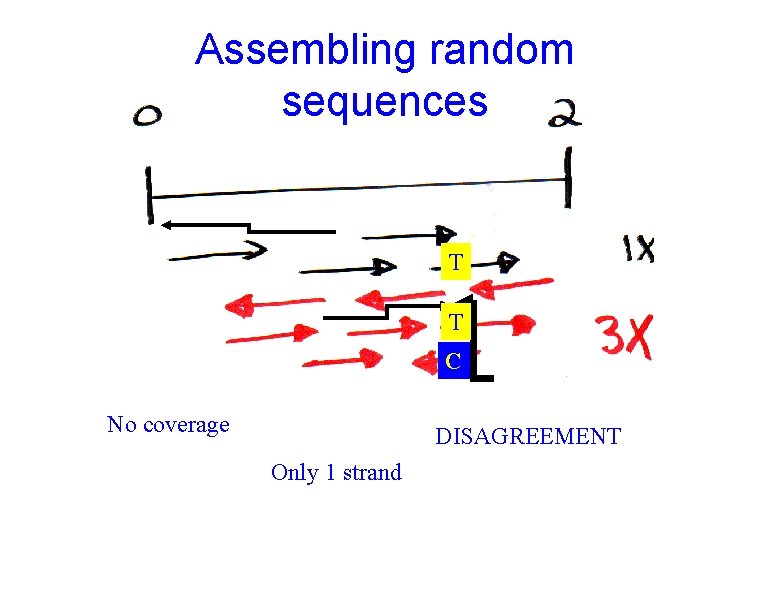
Assembling random sequences T T C No coverage DISAGREEMENT Only 1 strand
Assembly programs • Celera Assembler (Eugene Myers et al. ) • Arachne (Serafim Batzoglou et al. ) • PCAP (Xiaoqiu Huang, Iowa State University) • Phusion (EBI)

Continuing rapid improvement in sequencing technology
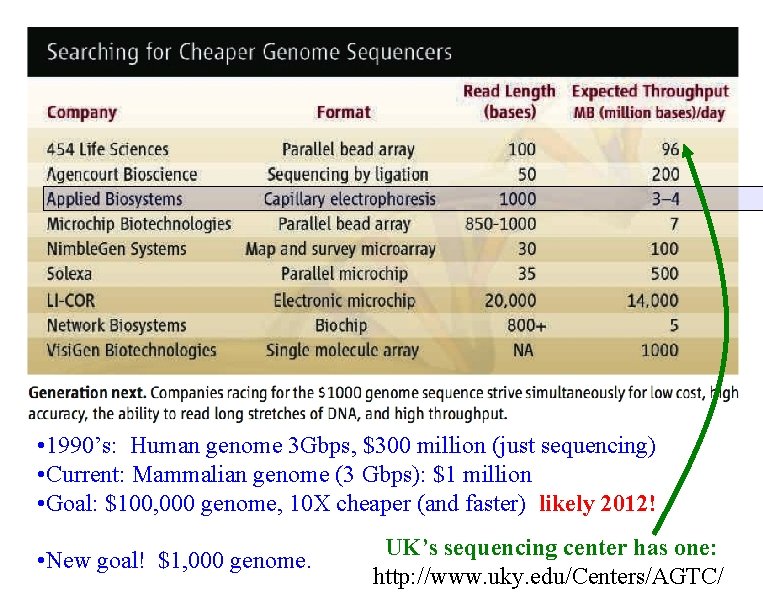
• 1990’s: Human genome 3 Gbps, $300 million (just sequencing) • Current: Mammalian genome (3 Gbps): $1 million • Goal: $100, 000 genome, 10 X cheaper (and faster) likely 2012! • New goal! $1, 000 genome. UK’s sequencing center has one: http: //www. uky. edu/Centers/AGTC/
454 Sequencing’s Genome Sequencer FLX • Pyrosequencing (sequencing by detection of nucleotides added during DNA synthesis. • 350 -400 million bases per run (10 hrs. ). • 400 bp sequence reads. • 1, 000 reads per run. • $6, 600 per run, 60 kb/$1, or $0. 00165/bp.
- Slides: 23