GENOM REGULATION BY LONG NONCODING RNA SUPERVISOR DR
GENOM REGULATION BY LONG NONCODING RNA SUPERVISOR: DR. FARAJOLLAHI PRESENTED BY: BAHAREH SADAT RASOULI
HOW BIG PART OF HUMAN TRANSCRIBED RNA RESULTS IN PROTEINS? • Of all RNA, transcribed in higher eukaryotes, 98% are never translated into proteins • Of those 98%, about 50 -70% are introns • The rest originate from non-protein genes, including r. RNA, t. RNA and a vast number of other non-coding RNAs (nc. RNAs) • Even introns have been shown to contain nc. RNAs, for example sno. RNAs

PROTEIN CODING GENES VS. BIOCOMPLEXITY
THREE MAIN CLASSES OF NCRNAS 1. Housekeeping RNA • t. RNA, r. RNA, sno. RNA, sn. RNA 2. Small regulatory RNA • mi. RNA (21 -24 nt), si. RNA 3. Long noncoding RNA (lnc. RNA) • ≥ 200 nt • Most prevalent class of nc. RNA • XIST, HOTAIR…
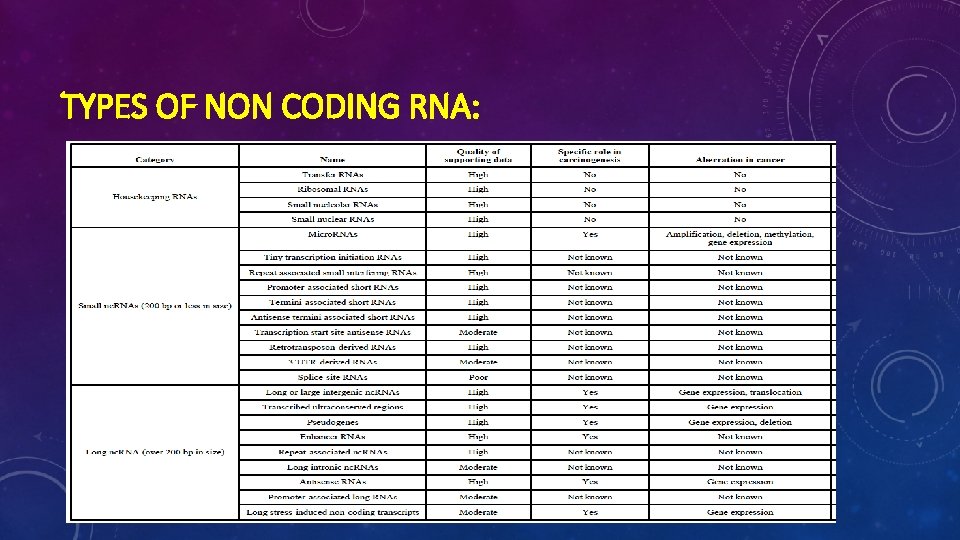
TYPES OF NON CODING RNA:

LONG NON CODING RNA: • There is no definition of lnc. RNA that is based on biological argumentation and widely accepted in the community. The most commonly used definition is based on the threshold of 200 nucleotides (nt) of the RNA length. • Lnc. RNAs are observed in a large diversity of species, including animals, plants, yeast, prokaryotes and even viruses. • lnc. RNAs are poorly conserved among different species when compared with the well-studied RNAs (such as m. RNAs, mi. RNAs, sno. RNAs)

LONG NON CODING RNA: • a considerable number of lnc. RNAs are 3' polyadenylated, 5' capped, multi-exonic and exhibit transcriptional activation activity similar to that of m. RNAs. • they seem mostly transcribed by RNA polymerase. II, but many long nc. RNAs avoid, or at least do not undergo, the subsequent standard m. RNA processing. • lnc. RNA promoters are bound and regulated by transcriptional factors, including Oct 3/4, Nanog, CREB, Sp 1, c-myc, Sox 2, NF-κB and p 53. • A feature intimately linked to their regulatory functions, which often entails nuclear retention close to the sites of transcription.

LONG NON CODING RNA: • Despite low expression of lnc. RNA, they play a significant role in a wide variety of important biological processes, including transcription, splicing, translation, protein localization, cellular structure integrity, imprinting, cell cycle and apoptosis, stem cell pluripotency and reprogramming and heat shock response. • It has been suggested that lnc. RNAs may regulate cancer progression and development of many other human diseases.

GENOMIC DISCOVERY OF LONG NONCODING RNAS • In addition to sequencing advances, new technologies were emerging to understand the regulation of gene expression and denovo identification of new genes. Ø Tiling microarray Ø RNA sequencing

TILING MICROARRAYS: • Tiling Arrays are a subtype of microarray chips. Like traditional microarrays, they function by hybridizing labeled DNA or RNA target molecules to probes fixed onto a solid surface. • Tiling arrays differ from traditional microarrays in the nature of the probes.

TILING MICROARRAYS PROBS:

TILING ARRAYS AID IN TRANSCRIPTOM MAPPING • discovering sites of DNA/protein interaction (Ch. IP-chip, Dam. ID), of DNA methylation (Me. DIPchip) and of sensitivity to DNase (DNase Chip). In addition to detecting previously unidentified genes and regulatory sequences, improved quantification of transcription products is possible

SYNTHESIS AND MANUFACTURERS • The two main ways of synthesizing tiling arrays are: Ø Photolitographic Ø Mechanical spotting/printing
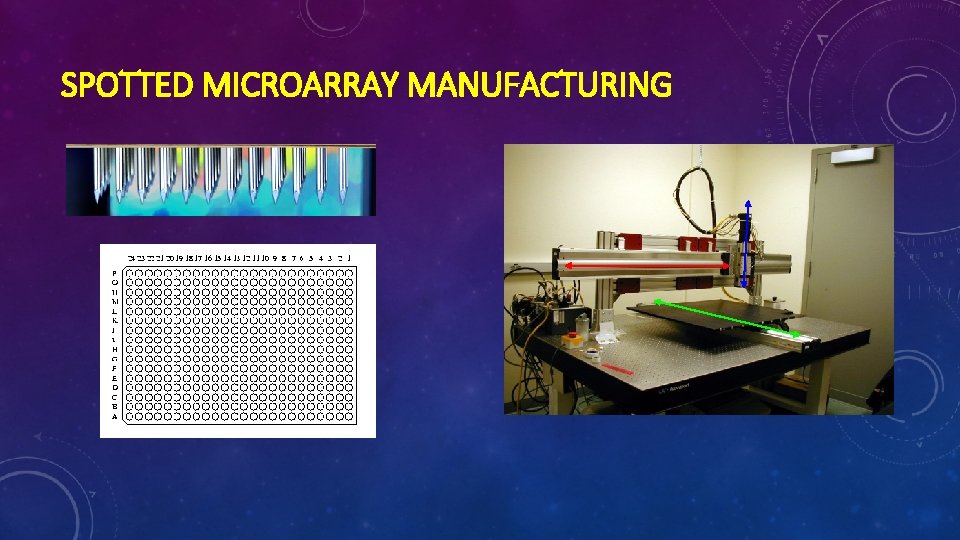
SPOTTED MICROARRAY MANUFACTURING

PHOTOLITOGRAPHIC:

RNA SEQUENCING: • RNA-seq (RNA Sequencing), also called "Whole Transcriptome Shotgun Sequencing" ("WTSS"), is a technology that uses the capabilities of next-generation sequencing to reveal a snapshot of RNA presence and quantity from a genome at a given moment in time.

RNA SEQUENCING:

RNA-SEQ VS. MICROARRAY • RNA-seq can be used to characterize novel transcripts and splicing variants as well as to profile the expression levels of known transcripts (but hybridization-based techniques are limited to detect transcripts corresponding to known genomic sequences) • RNA-seq has higher resolution than whole genome tiling array analysis • In principle, m. RNA can achieve single-base resolution, where the resolution of tiling array depends on the density of probes • RNA-seq can apply the same experimental protocol to various purposes, whereas specialized arrays need to be designed in these cases • Detecting single nucleotide polymorphisms (needs SNP array otherwise) • Mapping exon junctions (needs junction array otherwise) • Detecting gene fusions (needs gene fusion array otherwise) – Next-generation sequencing (NGS) technologies are now challenging microarrays as the tool of choice for genome analysis.

CLASSIFICATION OF LNCRNA ACCORDING TO GENOM LOCI: 1)Stand-alone lnc. RNAs 2)Natural antisense transcripts 3)Pseudogenes 4)Long intronic nc. RNAs 5) Divergent transcripts, promoter-associated transcripts, and enhancer RNAs
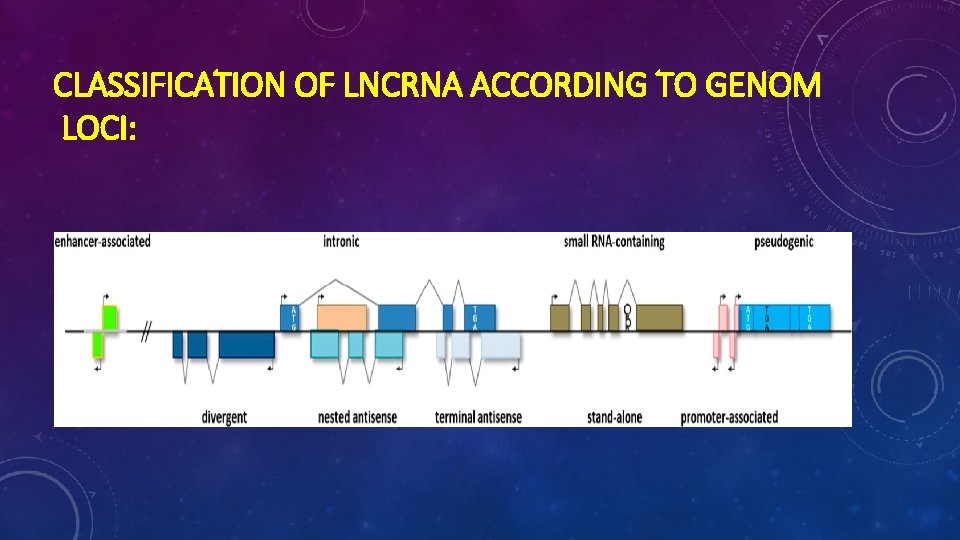
CLASSIFICATION OF LNCRNA ACCORDING TO GENOM LOCI:

FUNCTIONAL MECHANISMS OF LNCRNA: 1. Transcriptional regulation 2. Post – transcriptional regulation 3. Other function

TRANSCRIPTIONAL REGULATION: Effects Exerted on DNA Sequences • It has been found that lnc. RNAs are predominately localized in nucleus and chromatin, suggesting that lnc. RNAs may have a significant impact on DNA sequences. • Therefore, it is meaningful to classify lnc. RNAs based on their effects exerted on DNA sequences: Ø cis-lnc. RNAs Ø trans-lnc. RNAs v transcriptional interference and chromatin modification

TRANSCRIPTIONAL REGULATION:

POST TRANSCRIPTIONAL REGULATION: • There are two common post-transcriptional regulation mechanisms that lnc. RNAs get involved in, namely: Ø splicing regulation : function through binding or modulating splicing factors, or directly hybridizing with m. RNA sequences to block splicing. Ø translational control: function through binding to translation factors 27, 75 or ribosome Ø Natural antisense inhibitor: recent studies suggest that lnc. RNAs are also implicated in these processes, displaying direct si. RNA mechanisms or interacting with mi. RNAs.

POST TRANSCRIPTIONAL REGULATION:

OTHER MECHANISMS OF LNCRNA FUNCTIONING: Ø Protein localization Ø Telomeric replication Ø RNA interference Ø etc
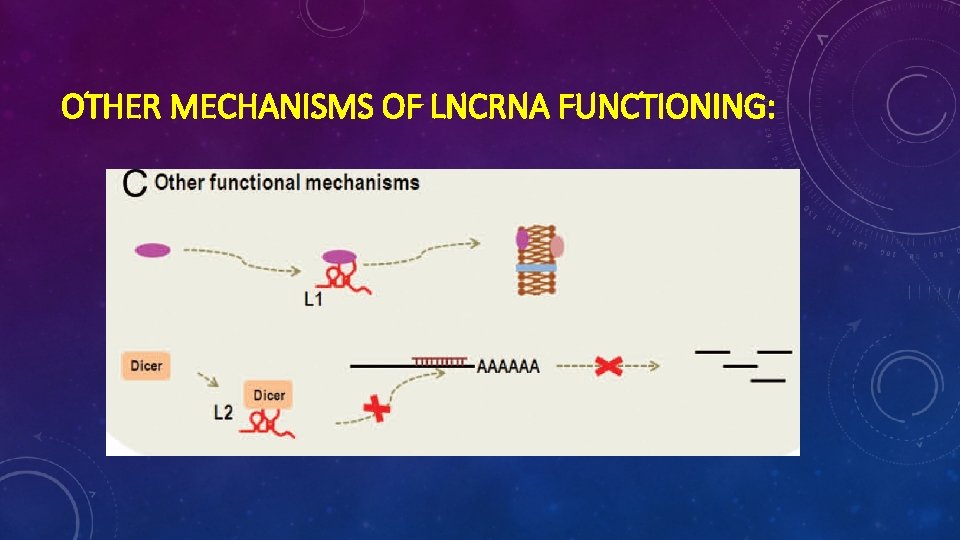
OTHER MECHANISMS OF LNCRNA FUNCTIONING:

TARGETING MECHANISMS OF LNCRNAS: • According to their mode of action, lnc. RNAs may also be classified based on their targeting mechanisms: Ø Signal : show cell type-specific expression and respond to diverse stimuli Ø Decoy : lnc. RNAs can serve as decoys that preclude the access of regulatory proteins to DNA Ø Guide : bind proteins and then direct the localization of ribonucleoprotein complex to specific targets Ø Scaffold : serve as central platforms to bring together multiple proteins to form ribonucleoprotein complexes

TARGETING MECHANISMS OF LNCRNAS:
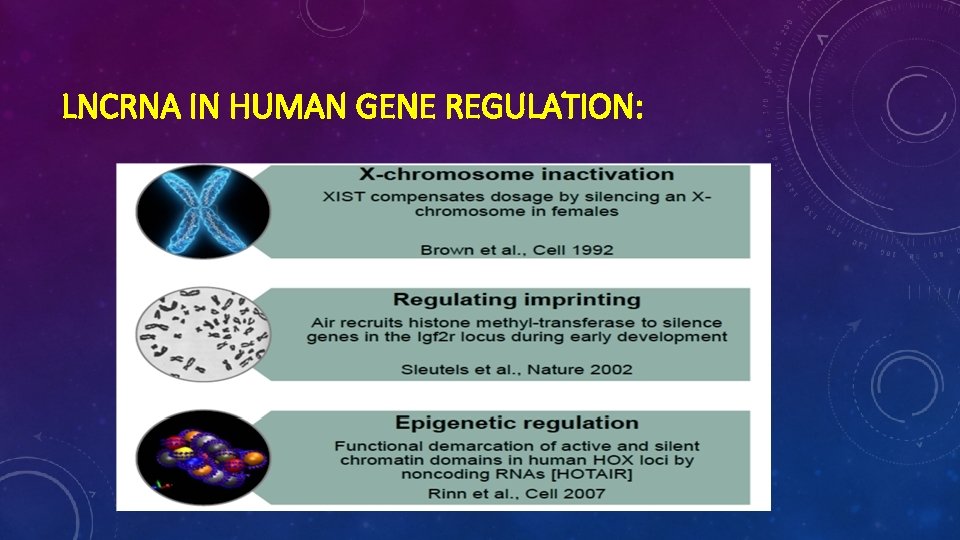
LNCRNA IN HUMAN GENE REGULATION:

X-CHROMOSOME DOSAGE COMPENSATION:

LNCRNA AND DISEASE: • Underscoring the importance of lnc. RNAs regulatory roles is their emergence as key players in etiology of several disease states. The strongest connection at present is with cancer. • lnc. RNA as disease markers

CONCLUSION: • The foregoing discussion has provided a survey of the present state of knowledge regarding the locations, functions, and mechanisms of lnc. RNAs. • A large fraction of the genome in many organisms is likely transcribed, but the characteristics and function of the overwhelming majority of these lnc. RNAs are currently not known. • A common emerging theme of lnc. RNA is that they form ribonucleic acis-protein interaction to carry out their function.

FUTURE ISSUES: • There is a need for large-scale LOF or gain-of-function studies to causally demonstrate lnc. RNA functions. • Genomic regions genetically associated with disease contain only lnc. RNAs, pointing to their genetic importance to disease, yet the functional roles of lnc. RNA remain largely unresolved. • There is a clear need to develop genetic model systems to understand lnc. RNAs’ function in vivo.

Thanks for your attention
- Slides: 35