GENETICS OF HEPATITIS B Dr Osama Hasan Othman

GENETICS OF HEPATITIS B Dr. Osama Hasan Othman MBCHB, DM, CABM FRCP LONDON KIRKUK MEDICAL COLLEGE KIRKUK IRAQ

KIRKUK MEDICAL COLLEGE

KIRKUK

GENETICS = SECTRETES OF LIFE
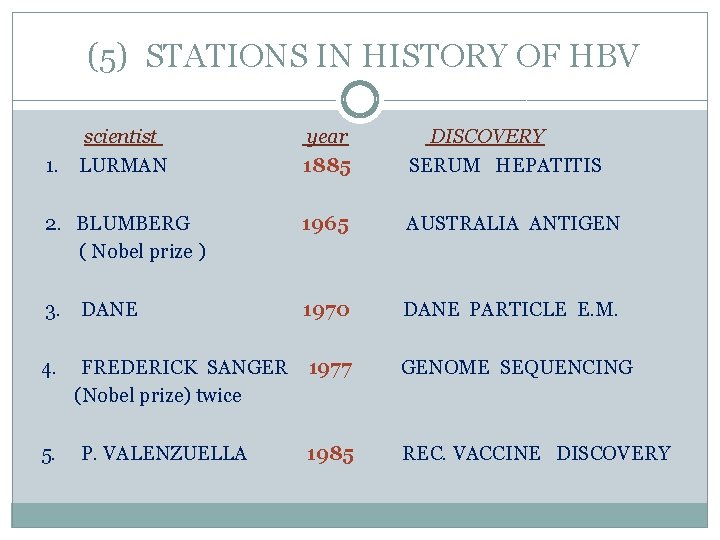
(5) STATIONS IN HISTORY OF HBV scientist year DISCOVERY 1. LURMAN 1885 SERUM HEPATITIS 2. BLUMBERG 1965 AUSTRALIA ANTIGEN ( Nobel prize ) 3. DANE 1970 DANE PARTICLE E. M. 4. FREDERICK SANGER 1977 GENOME SEQUENCING (Nobel prize) twice 5. P. VALENZUELLA 1985 REC. VACCINE DISCOVERY
• HBV GENOME STRUCTURE (5) features 1. The genome of HBV is made of circular DNA 2. It is unusual because the DNA is not fully doublestranded. 3. One end of the full length strand is linked to the viral DNA polymerase. 4. The genome is 3020– 3320 nucleotides long (for the full length strand) 5. While less 1700– 2800 nucleotides long (for the short length strand).

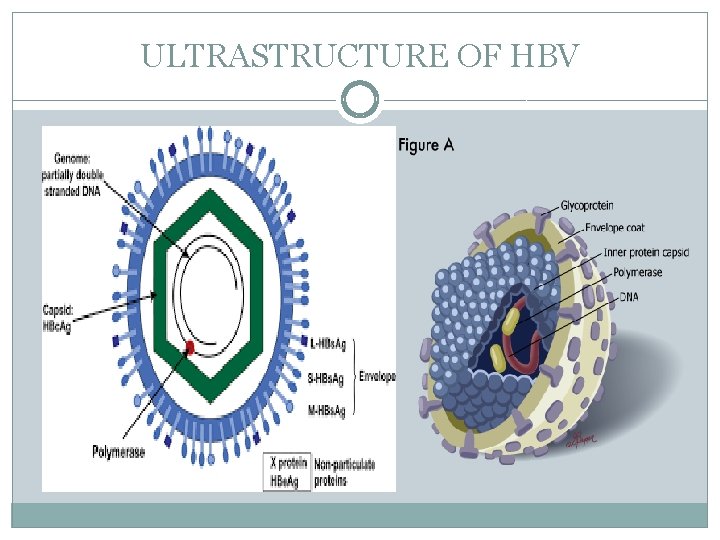
Unique features of HBV Although HBV small virion, many unique features make it distinctive among DNA Viruses: � It has a partially double stranded DNA with highly complex genome organization, life cycle and natural history. � It uses an RNA intermediate called pregenomic RNA (p g RNA) and Reverse Transcriptase RT for its genome replication. � Genome replication is accomplished by a complex mechanism of primer shifting facilitated by direct repeat sequences encoded in the genome. � The genome has evolved in such manner that every single nucleotide of the genome is used for either coding viral proteins or used as regulatory regions or both.
ULTRASTRUCTURE OF HBV
Sketch of HBV
HVB AS A CIRCLE
GENOME VARIABILITY � HVB utilizes internal in-frame translation initiation codons & different reading frames from the same RNA to generate different proteins with diverse functions. � HBV shows considerable genetic variability which has been related with clinical outcomes, replication potential, therapeutic responses.
COMPLETION OF VIRION � The negative-sense, (non-coding) strand is complementary to the viral m. RNA � The viral DNA is found in the nucleus soon after infection of the cell. �The partially double-stranded DNA is rendered fully double-stranded by completion of the (+) sense strand by viral polymerase and removal of a protein molecule from the (-) sense strand a short sequence of RNA from the (+) sense strand. � Non-coding bases are removed from the ends of the (-) sense strand the ends are rejoined.
VIRAL TRANSCRIPTION �The viral genes are transcribed by the cellular RNA polymerase II in the cell nucleus from a covalently closed circular DNA (ccc DNA) template. �Two enhancers designated enhancer I (Enh I) and enhancer II (Enh II) have been identified. Both enhancers exhibit greater activity in cells of hepatic origin, they drive and regulate the expression of the complete viral transcripts.
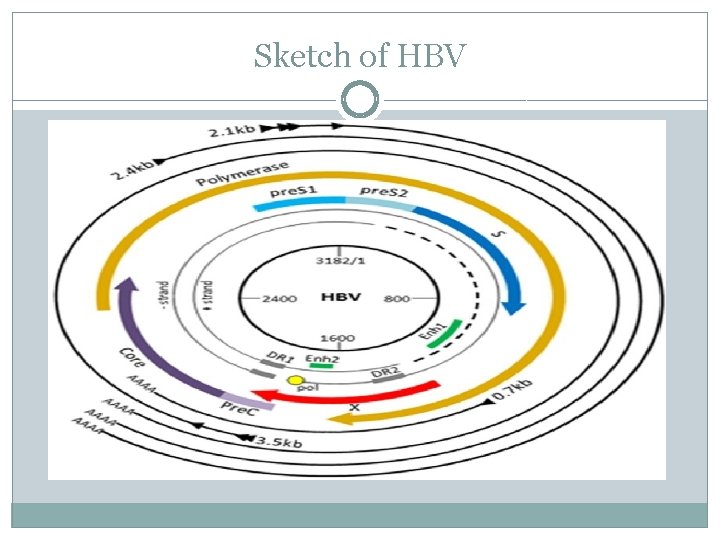
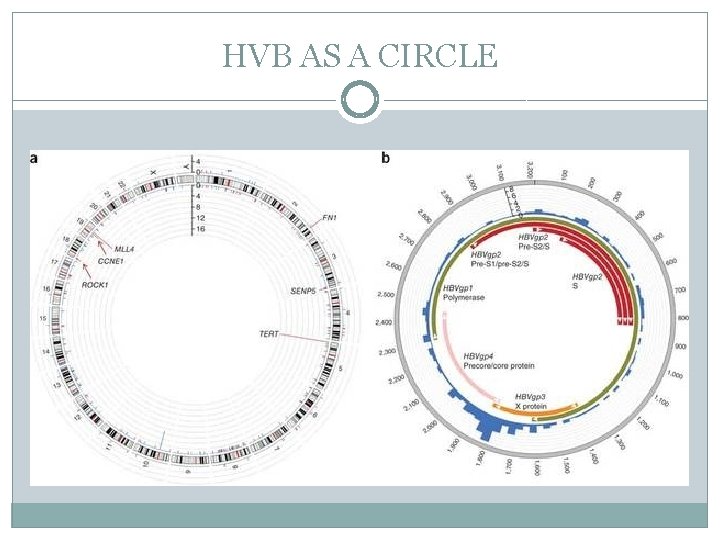
GENES OF GENOME Four genes encoded by genome called C, P, S, and X. The core protein is coded for by gene C (HBc. Ag), its start codon preceded by an AUG start codon from which the pre-core protein produced. HBe. Ag produced by proteolytic processing of pre-core protein The DNA polymerase is encoded by gene P. Gene S , the gene that codes for the surface antigen (HBs. Ag). The HBs. Ag gene has one long open reading frame, contains three in frame "start" (ATG) codons that divide the gene into three sections, pre-S 1, pre. S 2, and S. Because of multiple start codons, polypeptides of three different sizes called large, middle, and small (p re-S 1 + pre-S 2 + S, or S) produced. The function of the protein coded for by gene X is not fully understood, but evidence suggests it may function as transcriptional trans activator. Several non-coding RNA elements have been identified in the HBV genome. These include: HBV PRE alpha, HBV PRE beta and HBV RNA encapsidation signal epsilon

GENOTYPES & SUBTYPES Genotypes differ by at least 10% of sequence and have distinct geographical distributions , has been associated with anthropological history. � Within genotypes subtypes have been described: these differ by 5% of the genome. � There are eight known genotypes labeled A through H. � A possible new "I" genotype has been described Two further genotypes have since been recognised. The current (2014) listing now runs A though to J , so (10) genotypes � Several subtypes also recognized, there are (24) subtypes. Different genotypes may respond to treatment in different ways

Geographical distribution of Individual genotypes area: blue sky! � Type F which diverges from other genomes by 14% the most divergent type known. � Type A is prevalent in Europe, Africa and South-east Asia, including the Philippines. � Type B and C are predominant in Asia. � type D is common in the Mediterranean area, the Middle East and India. � type E is localized in sub-Saharan Africa. � type F (or H) is restricted to Central and South America. � Type G has been found in France and Germany. � Genotypes A, D and F are predominant in Brazil and all genotypes occur in the United States with frequencies dependent on ethnicity. � The E and F strains appear to have originated in aboriginal populations of Africa and the New World, respectively
Further geographical divisions � Type B hans two distinct geographical distributions: Bj/B 1 ('j'—Japan) and Ba/B 2 ('a'—Asia). Type Ba has been further subdivided into four clades (B 2–B 4). � Type C has two geographically subtypes: Cs (C 1) in Southeast Asia and C e (C 2) in East Asia. The C subtypes have been divided into five clades (C 1–C 5). A sixth clade (C 6) has been described in the Philippines but only in one isolate to date. � Type C 1 is associated with Vietnam, Myanmar and Thailand; � type C 2 with Japan, Korea and China; � type C 3 with New Caledonia and Polynesia; � C 4 with Australia; � and C 5 with the Philippines. A further subtype has been described in Papua, Indonesia.
subdivisions �Type D has been divided into 7 subtypes (D 1–D 7). �Type F has been subdivided into 4 subtypes (F 1–F 4). F 1 has been further divided into 1 a and 1 b. In Venezuela subtypes F 1, F 2, and F 3 are found in East and West Amerindians. �Among South Amerindians only F 3 was found. Subtypes I a, III, and IV exhibit a restricted geographic distribution (Central America, the North and the South of South America respectively) while clades I b and II are found in all the Americas except in the Northern South America and North America respectively.
Life cycle of HBV � The life cycle of hepatitis B virus is complex. Hepatitis B is one of a few known non-retroviral viruses which use reverse transcription as a part of its replication process. 1. Attachment: The virus gains entry into the cell by binding to a receptor on the surface of the cell and enters it by clathrin-dependent endocytosis. The cell surface receptor has been identified as the Sodium/Bile acid cotransporting pepetide SLC 10 A 1 (also named NTCP). 2. Penetration: The virus membrane then fuses with the host cell's membrane releasing the DNA and core proteins into the cytoplasm.
LIFE CYCLE OF HBV 3. Uncoating: the virus multiplies via RNA made by host enzyme, the viral genomic DNA transferred to the cell nucleus. The capsid transported to microtubules via nuclear pore. The core proteins dissociate from the partially double stranded viral DNA & fully double stranded DNA formed and transformed into covalently closed circular DNA (ccc. DNA) that serves as a template for transcription of four viral m. RNAs. 4. Replication: The largest m. RNA, ( longer than the viral genome), used to make the new copies of the genome and make the capsid core protein viral DNA polymerase. 5. Assembly: These four viral transcripts undergo additional processing and form progeny virions then released from the cell or returned to the nucleus and re-cycled to produce even more copies. 6. Release: The long m. RNA then transported back to the cytoplasm where virion P protein synthesizes DNA via its reverse transcriptase activity.

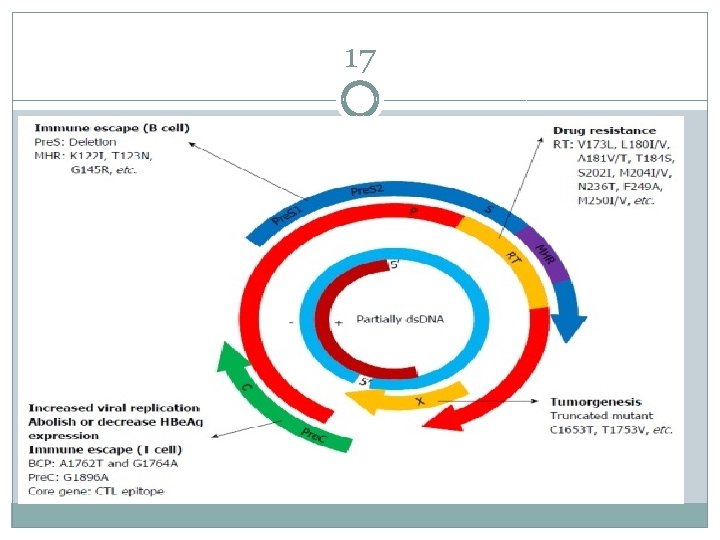
Genome Diversities Related to Clinical Outcome � Naturally occurring mutations on HBV genome are located at the CORF. The dual BCP mutation A 1762 T + G 1764 A results in a down regulation of HBe. Ag synthesis, and the PC G 1896 A stop-codon mutation prevents the expression of HBe. Ag. � Another PC mutation A 1899 G has been reported in combination with G 1896 A, which stabilizes the lower stem of the “ϵ” encapsidation signal and enhances the replication. The presence of both BCP and PC variants has been reported in severe liver cirrhosis and HCC. � Several mutations, including deletions and insertions or stop codons have been reported in the SORF of the HBV genome.
CLINICAL OUTCOME OF MUTATIONS Deletions or missense mutations in Pre-S 2 region can abolish the synthesis of the protein and alter B and T cell epitopes. HBs. Ag insertion or deletion or missense mutations can help the virus to evade host immune response. Truncated HBV surface proteins contribute to chronicity of HBV. a pattern of hotspot mutations located at the X region of the HBV genome (x. I 127 T, x. K 130 M, x. V 131 I and x. F 132 Y) are known to be associated with transactivating function and HCC development.
Genome Variations Related to FHF �It has been hypothesized that both viral and host factors play a role in the pathogenesis and clinical outcome of HBV infection. � FHF develops when there is an overwhelming immune-mediated lysis of infected hepatocytes It has been suggested that FHB can be explained by three main virologic markers : �(i) increase in viral replication fitness. �(ii) change in viral gene expression. �(iii) alteration of B and T cell epitopes.
mutations in overlapping envelope & polymerase genes of HBV � 1. The potential for (HBV) to alter its genome is considerable. � 2. This potential occurs as the virus utilizes a reverse transcription step in replicating it s genome. � 3. Like HIV virus, the reverse transcriptase of HBV is error prone and as a consequence of specific selection pressures within a host a population of viral quasi species emerges. � 4. HBV mutants with survival advantages over the wild type virus appear within the selective in vivo environment. � 5. Some of these viruses include HBV vaccine escape and anti-viral resistant mutants that have changes in the envelope (S) and polymerase genes, respectively
21
17
Pre core & core GENE VARIATION & FHB Many evidences suggest that FHB strongly associated with HBV strains not producing HBe. Ag due to BCP or PC mutations. HBV DNA clones were propagated from FHB patients and sequenced within the BCP and PC region. Interestingly, a significant number of clones carried the PC G 1896 A stop-codon and G 1899 A mutations & dual BCP A 1762 T + G 1764 A mutation. The precursor protein of HBe. Ag decreases the encapsidation of the pregenomic RNA.
Sequences of mutations Absence or decrease of HBe. Ag synthesis can lead to enhanced viral replication and consequently increased host immune response The double A 1762 T + G 1764 A mutation can slightly increase viral DNA replication The C 1766 T + T 1768 A mutational pattern exhibits a 10 -fold higher replication capacity than a wild type strain. Mutations can synergistically influence HBV replication. For instance, BCP and PC can further enhance the replication of G 145 R and other“a-determinant” mutants

Overlapping mutations The genome of HBV is organized in to overlapping reading frames. The S gene is completely overlapped by the polymerase gene. As a consequence, mutations in the gene may produce changes in the overlapping polymerase gene. mutations in the polymerase gene may produce changes in the S gene. Certain mutations in either the S or polymerase gene produce functionally significant changes in the respective overlapping gene. Treatment of chronic hepatitis B carriers with long-term lamivudine (LMV) results in the selection of HBV mutants that are resistant to this nucleoside analogue.
LMV resistance & changes in Genome The polymerase mutations associated with LMV resistance produce changes in the overlapping S gene and in its envelope protein ( HBs. Ag) that results in a reduced antigenicity of the HBs. Ag protein. The selection of vaccine escape mutants by HBV vaccination or hepatitis B immune globulin is associated with changes in the S gene that are accompanied by mutations in the fingers sub-domain of the polymerase protein. When combined with polymerase mutations that are associated with resistance to LMV the changes within the fingers sub-domain of the viral enzyme behave as compensatory mutations that are able to restore the replication of LMV resistant HBV. The ability to change a viral protein by mutations in an overlapping but unrelated viral gene may produce HBV mutants with altered antigenicity and/or replication and a natural history that may be distinctly different to wild type HBV.
Genotypes & pathogenesis + outcome � As HBV genotypes and sub-genotypes have distinct geographical distributions, abundant evidence has shown that different genotypes and sub-genotypes are associated with distinctive pathogenesis and outcomes of HBV infection. � Generally, HBV genotype C has been associated with an increased risk of liver inflammation, flares of hepatitis, liver fibrosis and HCC. � Genotypes D, C, and F 1 are more likely to develop complications such as liver cirrhosis and HCC. � Hbe. Ag seroconversion occurred much later in patients infected with genotype C compared to other genotypes.
MUTATIONS IN SPECIFIC SITES Mutations in the pre S/S region are associated with: 1. vaccine failure 2. immune escape 3. occult HBV infection 4. HCC Mutations in the P region may cause drug resistance to NA antivirals. Mutations in the pre. C/C region are related to HBe. Ag negativity, immune escape, and persistent hepatitis. Mutations in the X region play critical roles in promoting HCC.
Clinical significance of mutations Surface gene mutations were initially noted as vaccine escape mutants in HBV endemic regions receiving HBV immuno prophylaxis at birth. These mutations have subsequently been noted in patients after liver transplantation who exhibit HBV recurrence despite receiving hepatitis B immune globulin ( immune escape mutant). Surface gene mutations in the pre-S 2 region have been associated with fulminant hepatitis
Mutation & disease activity Mutations in the precore, and surface region have been associated with a broad range of disease activity, varying from asymptomatic HBV carriage to fulminant hepatitis. Mutations in the basal core promotor have been associated in vitro with enhanced viral replication. Detailed genomic studies have sought to correlate distinct mutations with enhanced virulence, however, it remains difficult to dissect the role of viral versus host factors in this issue.
Polymerase variants & Antivirals A variety of polymerase mutations have been shown after HBV therapy with antiviral nucleosides inhibitors. Treatment with either lamivudine or famciclovir has been associated with the development of polymerase variants in HBV isolates. Most patients developing polymerase YMDD variants continue to derive clinical benefit from prolonged lamivudine therapy, which may be secondary to the diminished viral replication conferred by these nucleotide changes. Patients on extended lamivudine therapy harboring these variants may be identified by the presence of persistent HBe. Ag and moderate elevations of ALT and DNA. .

THANK YOU Dr. Gene. .
- Slides: 36