Genetic Recombination Definition The breakage and joining of
Genetic Recombination Definition: The breakage and joining of DNA into new combinations Purposes • Promotes genetic diversity within a species - within a chromosome causes inversions, deletions, duplications - horizontal exchange introduces new sequences (information) • Plays a major role in repair of damaged DNA and mutagenesis • Critical for several mechanisms of phase and antigenic variation In the lab: introduce foreign DNA or mutations into bacteria map the distance between mutations

Types: • Homologous recombination (or general recombination) • basic steps • current models • proteins that play a role • practical applications • Nonhomologous recombination (site-specific) • Basic steps • general categories of proteins used • examples – phage integration, flagellin phase variation • Illegitimate recombination (transposition)
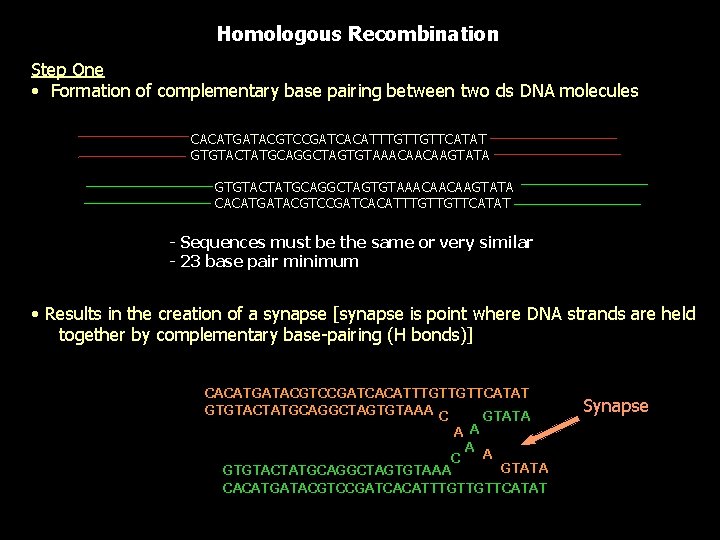
Homologous Recombination Step One • Formation of complementary base pairing between two ds DNA molecules CACATGATACGTCCGATCACATTTGTTGTTCATAT GTGTACTATGCAGGCTAGTGTAAACAACAAGTATA CACATGATACGTCCGATCACATTTGTTGTTCATAT - Sequences must be the same or very similar - 23 base pair minimum • Results in the creation of a synapse [synapse is point where DNA strands are held together by complementary base-pairing (H bonds)] CACATGATACGTCCGATCACATTTGTTGTTCATAT GTGTACTATGCAGGCTAGTGTAAA C GTATA A A C GTATA GTGTACTATGCAGGCTAGTGTAAA CACATGATACGTCCGATCACATTTGTTGTTCATAT Synapse
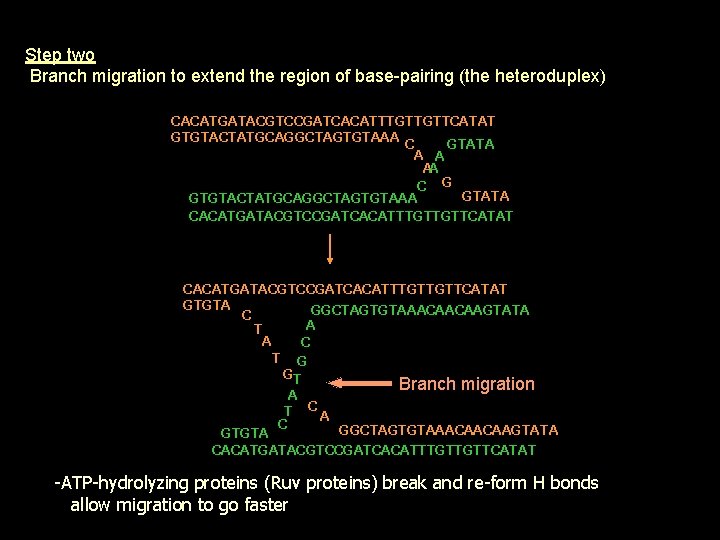
Step two Branch migration to extend the region of base-pairing (the heteroduplex) CACATGATACGTCCGATCACATTTGTTGTTCATAT GTGTACTATGCAGGCTAGTGTAAA C GTATA A A AA C G GTATA GTGTACTATGCAGGCTAGTGTAAA CACATGATACGTCCGATCACATTTGTTGTTCATAT GTGTA GGCTAGTGTAAACAACAAGTATA C A T A C T G GT Branch migration A T CA C GGCTAGTGTAAACAACAAGTATA GTGTA CACATGATACGTCCGATCACATTTGTTGTTCATAT -ATP-hydrolyzing proteins (Ruv proteins) break and re-form H bonds allow migration to go faster
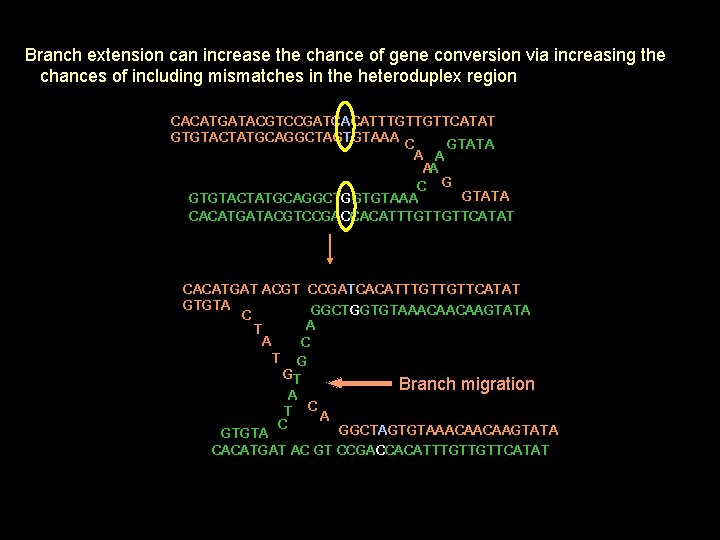
Branch extension can increase the chance of gene conversion via increasing the chances of including mismatches in the heteroduplex region CACATGATACGTCCGATCACATTTGTTGTTCATAT GTGTACTATGCAGGCTAGTGTAAA C GTATA A A AA C G GTATA GTGTACTATGCAGGCTGGTGTAAA CACATGATACGTCCGACCACATTTGTTGTTCATAT CACATGAT ACGT CCGATCACATTTGTTGTTCATAT GTGTA GGCTGGTGTAAACAACAAGTATA C A T A C T G GT Branch migration A T CA C GGCTAGTGTAAACAACAAGTATA GTGTA CACATGAT AC GT CCGACCACATTTGTTGTTCATAT

Step three Resolution of the heteroduplex - isomerization of the duplex due to strands uncrossing and re-crossing - results in different outcomes upon resolution - 50% chance of each isomer being resolved
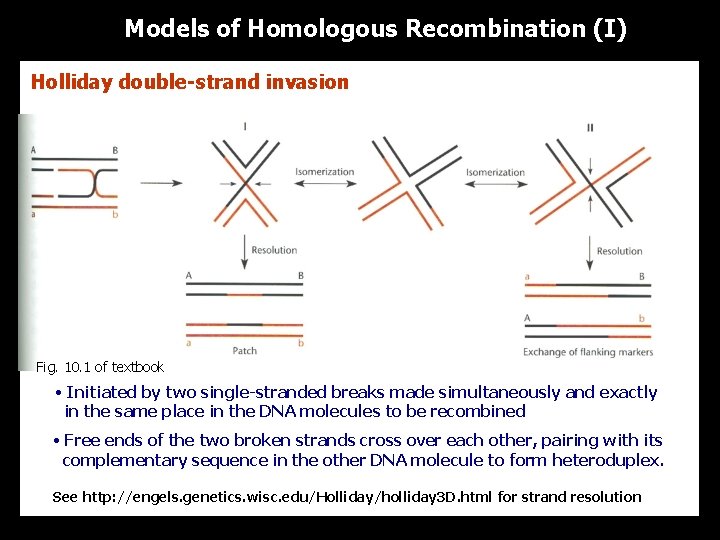
Models of Homologous Recombination (I) Holliday double-strand invasion Fig. 10. 1 of textbook • Initiated by two single-stranded breaks made simultaneously and exactly in the same place in the DNA molecules to be recombined • Free ends of the two broken strands cross over each other, pairing with its complementary sequence in the other DNA molecule to form heteroduplex. See http: //engels. genetics. wisc. edu/Holliday/holliday 3 D. html for strand resolution
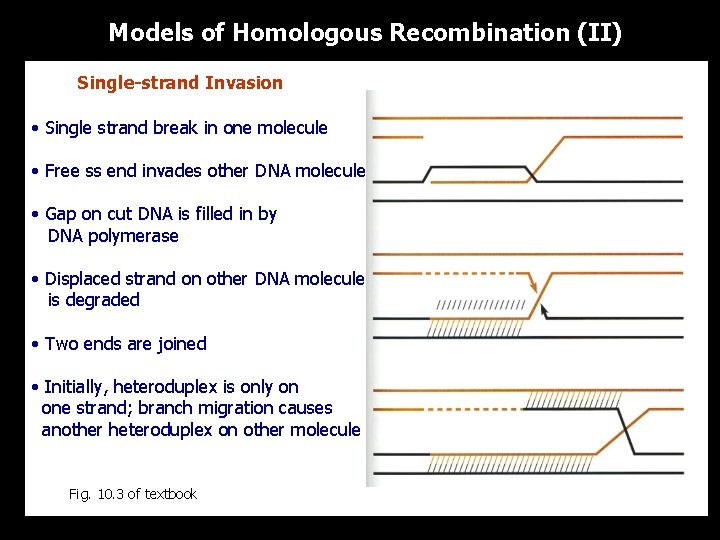
Models of Homologous Recombination (II) Single-strand Invasion • Single strand break in one molecule • Free ss end invades other DNA molecule • Gap on cut DNA is filled in by DNA polymerase • Displaced strand on other DNA molecule is degraded • Two ends are joined • Initially, heteroduplex is only on one strand; branch migration causes another heteroduplex on other molecule Fig. 10. 3 of textbook
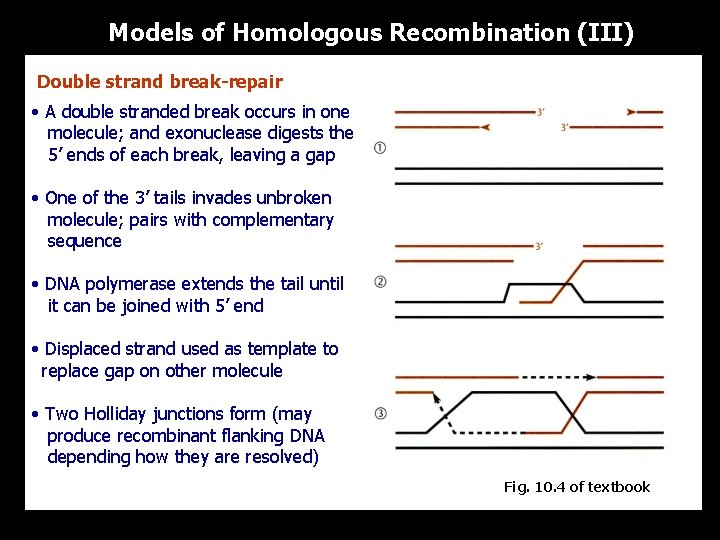
Models of Homologous Recombination (III) Double strand break-repair • A double stranded break occurs in one molecule; and exonuclease digests the 5’ ends of each break, leaving a gap • One of the 3’ tails invades unbroken molecule; pairs with complementary sequence • DNA polymerase extends the tail until it can be joined with 5’ end • Displaced strand used as template to replace gap on other molecule • Two Holliday junctions form (may produce recombinant flanking DNA depending how they are resolved) Fig. 10. 4 of textbook

Proteins involved in DNA recombination (the E. coli paradigm) Mutation Phenotype Rec. A Recombination deficient Rec. BC Reduced recombination Rec. D Rec+ Rec. F Reduced plasmid recombination Rec. J Rec. O Rec. R Rec. Q Reduced recombination in Rec. BCas above independent Rec. N Rec. G Reduced recombination in Rec. BCReduced recombination in Ruv. A-B-C- Ruv. A Ruv. B Ruv. C Reduced recombination in Rec. Gas above Pri. A Pri. B Pri. C Dna. T Reduced recombination as above + or Donor DNA Mutant bank (i. e. of E. coli) Screen for inability to acquire a selectable marker
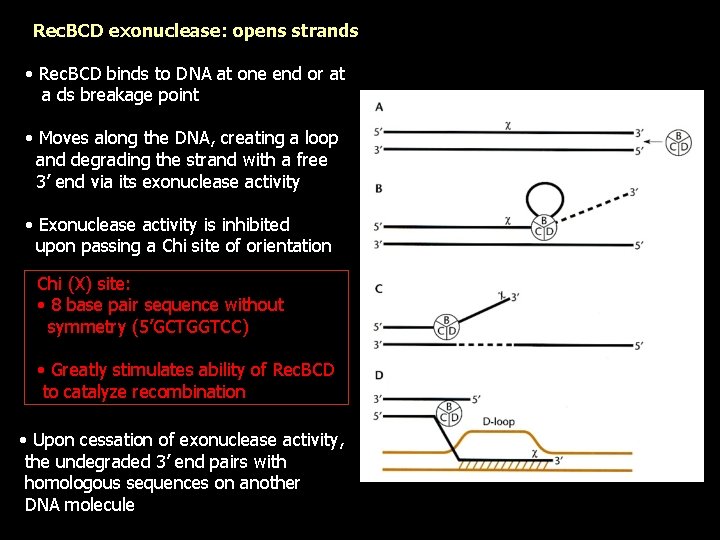
Rec. BCD exonuclease: opens strands • Rec. BCD binds to DNA at one end or at a ds breakage point • Moves along the DNA, creating a loop and degrading the strand with a free 3’ end via its exonuclease activity • Exonuclease activity is inhibited upon passing a Chi site of orientation Chi (Χ) site: • 8 base pair sequence without symmetry (5’GCTGGTCC) • Greatly stimulates ability of Rec. BCD to catalyze recombination • Upon cessation of exonuclease activity, the undegraded 3’ end pairs with homologous sequences on another DNA molecule
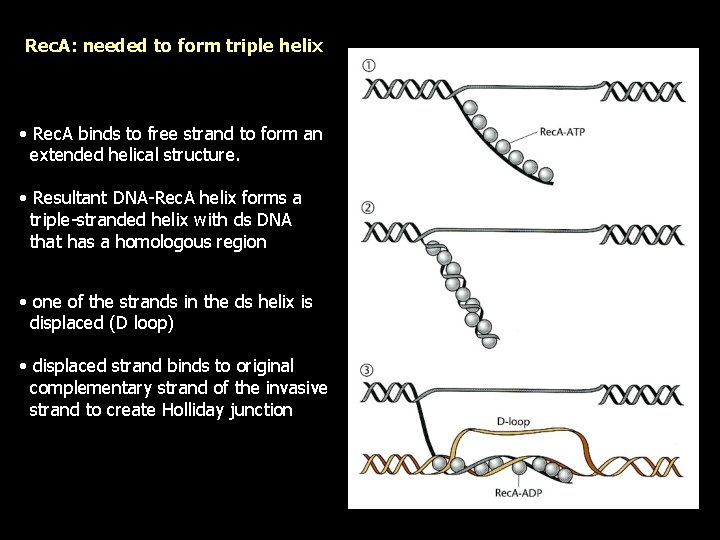
Rec. A: needed to form triple helix • Rec. A binds to free strand to form an extended helical structure. • Resultant DNA-Rec. A helix forms a triple-stranded helix with ds DNA that has a homologous region • one of the strands in the ds helix is displaced (D loop) • displaced strand binds to original complementary strand of the invasive strand to create Holliday junction

Rec. A protein-ds. DNA complex imaged by atomic force microscopy www-mic. ucdavis. edu
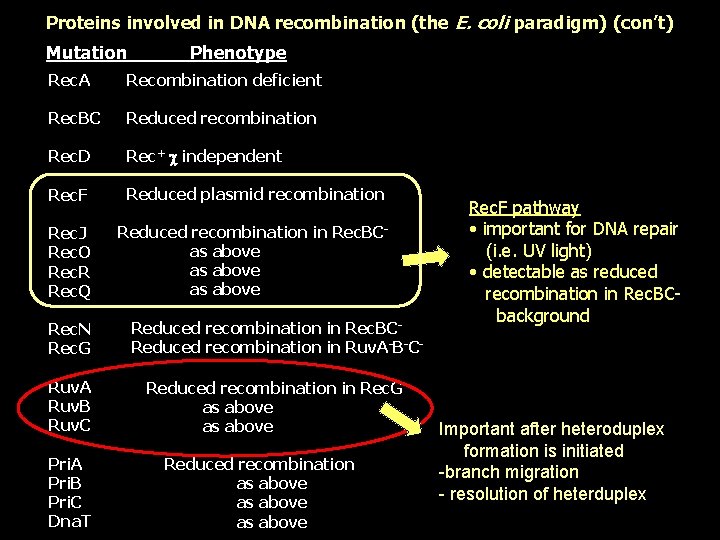
Proteins involved in DNA recombination (the E. coli paradigm) (con’t) Mutation Phenotype Rec. A Recombination deficient Rec. BC Reduced recombination Rec. D Rec+ independent Rec. F Reduced plasmid recombination Rec. J Rec. O Rec. R Rec. Q Reduced recombination in Rec. BCas above Rec. N Rec. G Reduced recombination in Rec. BCReduced recombination in Ruv. A-B-C- Ruv. A Ruv. B Ruv. C Reduced recombination in Rec. Gas above Pri. A Pri. B Pri. C Dna. T Reduced recombination as above Rec. F pathway • important for DNA repair (i. e. UV light) • detectable as reduced recombination in Rec. BCbackground Important after heteroduplex formation is initiated -branch migration - resolution of heterduplex

Efficient branch migration requires Ruv. A and Ruv. B • Ruv. A specifically binds Holliday junctions - resultant structure better able to undergo branch migration and resolution • Ruv. B is a helicase - forms a hexameric ring around the DNA strand - DNA is pumped through the ring using ATP cleavage to drive the pump - the synapse is thus forced to migrate Ruv. B Ruv. A Ruv. C resolves (cuts) the Holliday junction • Ruv C is a specialized endonuclease an X-phile – cuts crossed DNA strands always cuts at two T’s
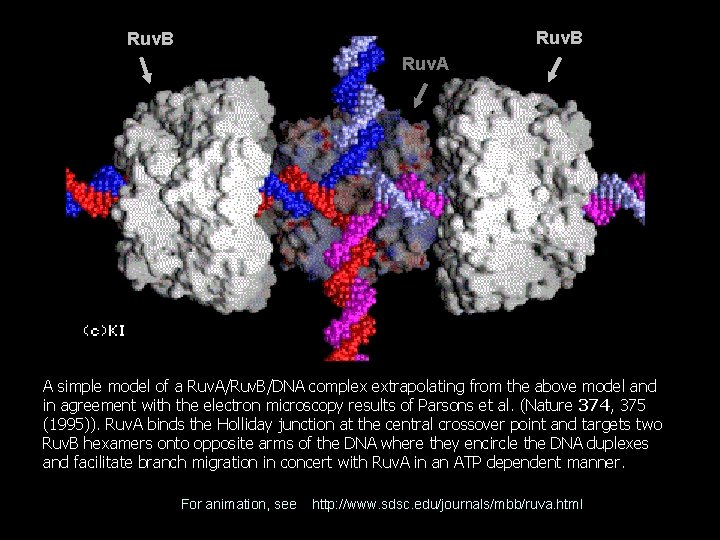
Ruv. B Ruv. A A simple model of a Ruv. A/Ruv. B/DNA complex extrapolating from the above model and in agreement with the electron microscopy results of Parsons et al. (Nature 374, 375 (1995)). Ruv. A binds the Holliday junction at the central crossover point and targets two Ruv. B hexamers onto opposite arms of the DNA where they encircle the DNA duplexes and facilitate branch migration in concert with Ruv. A in an ATP dependent manner. For animation, see http: //www. sdsc. edu/journals/mbb/ruva. html

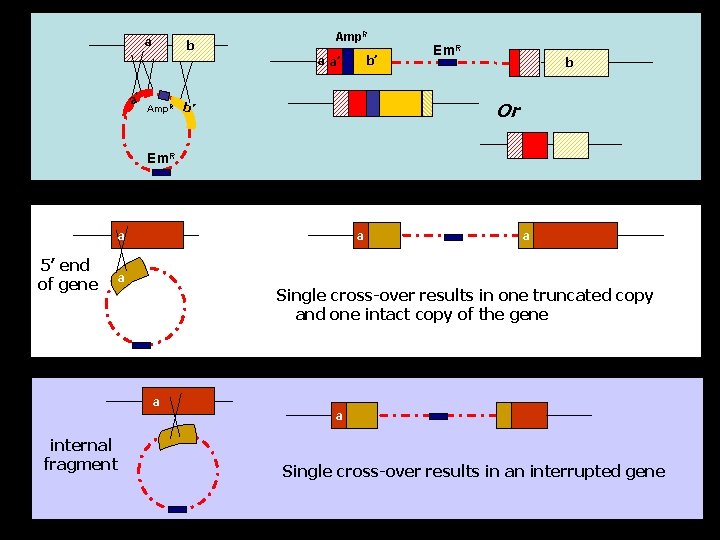
a b Amp. R b’ a a’ a’ Em. R b Or Amp. R b’ Em. R a 5’ end of gene a a a Single cross-over results in one truncated copy and one intact copy of the gene a internal fragment a Single cross-over results in an interrupted gene
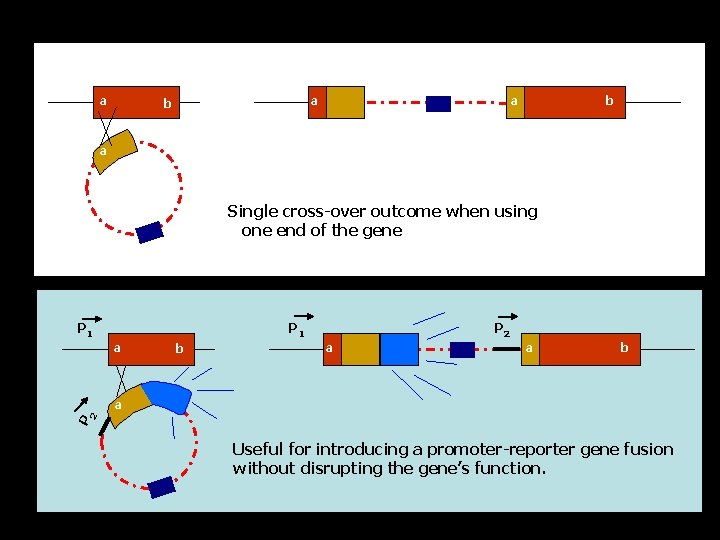
a a a b b a Single cross-over outcome when using one end of the gene P 1 a b P 2 a Useful for introducing a promoter-reporter gene fusion without disrupting the gene’s function.

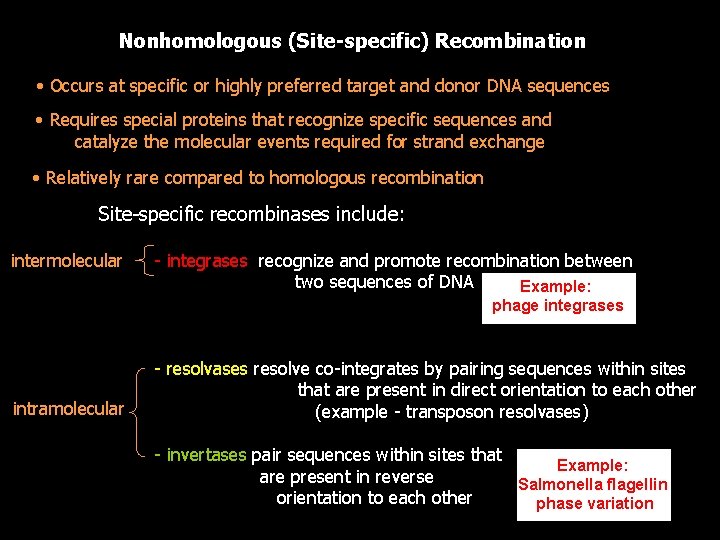
Nonhomologous (Site-specific) Recombination • Occurs at specific or highly preferred target and donor DNA sequences • Requires special proteins that recognize specific sequences and catalyze the molecular events required for strand exchange • Relatively rare compared to homologous recombination Site-specific recombinases include: intermolecular - integrases recognize and promote recombination between two sequences of DNA Example: phage integrases intramolecular - resolvases resolve co-integrates by pairing sequences within sites that are present in direct orientation to each other (example - transposon resolvases) - invertases pair sequences within sites that Example: are present in reverse Salmonella flagellin orientation to each other phase variation
Lytic/Lysogenic Developmental Switch
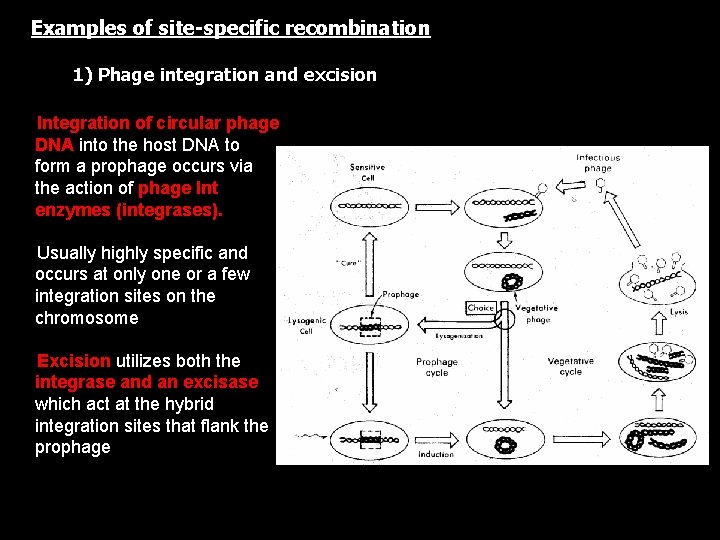
Examples of site-specific recombination 1) Phage integration and excision • Integration of circular phage DNA into the host DNA to form a prophage occurs via the action of phage Int enzymes (integrases). • Usually highly specific and occurs at only one or a few integration sites on the chromosome • Excision utilizes both the integrase and an excisase, which act at the hybrid integration sites that flank the prophage
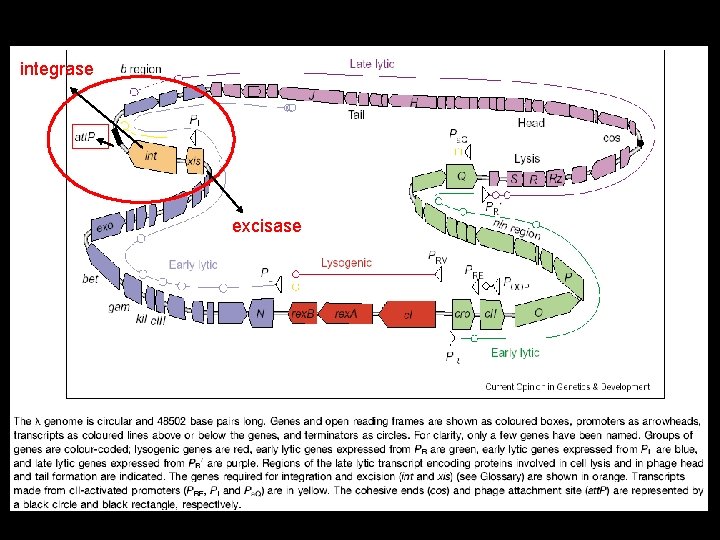
integrase excisase
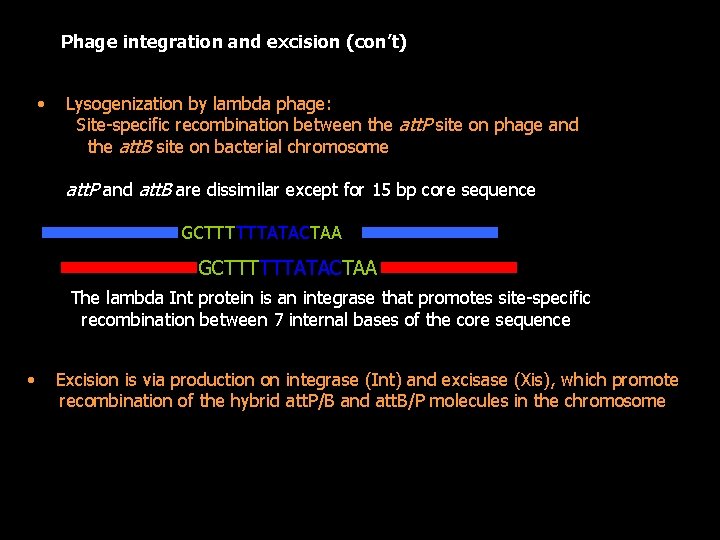
Phage integration and excision (con’t) • Lysogenization by lambda phage: Site-specific recombination between the att. P site on phage and the att. B site on bacterial chromosome att. P and att. B are dissimilar except for 15 bp core sequence GCTTTTTTATACTAA The lambda Int protein is an integrase that promotes site-specific recombination between 7 internal bases of the core sequence • Excision is via production on integrase (Int) and excisase (Xis), which promote recombination of the hybrid att. P/B and att. B/P molecules in the chromosome
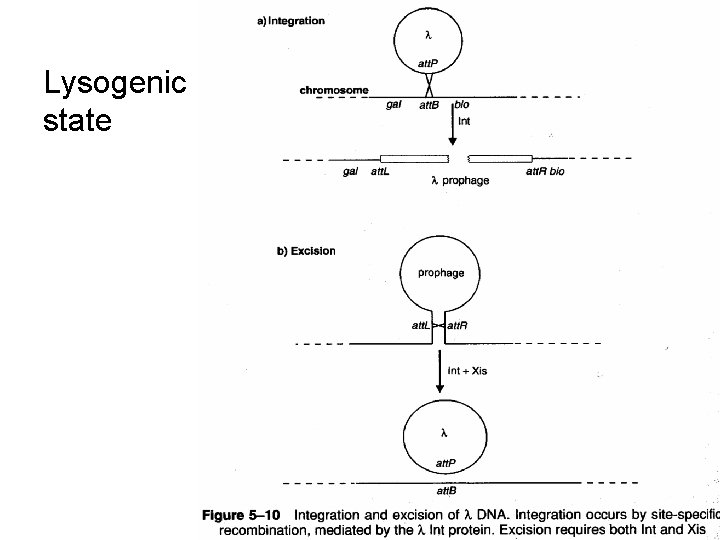
Lysogenic state
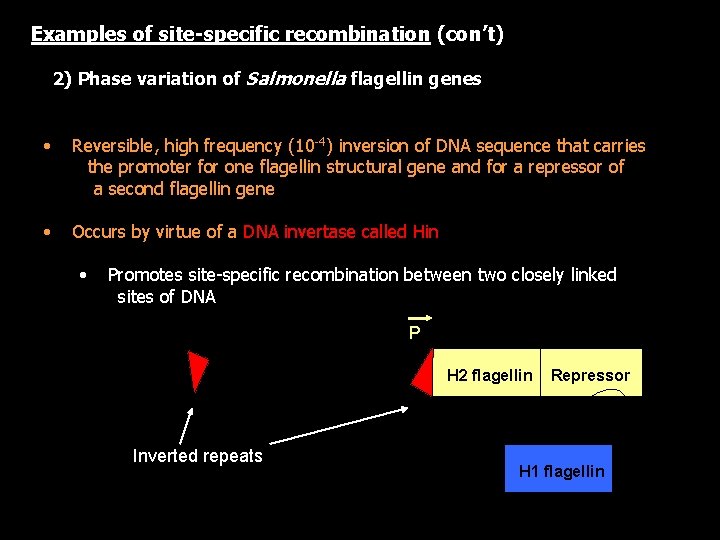
Examples of site-specific recombination (con’t) 2) Phase variation of Salmonella flagellin genes • Reversible, high frequency (10 -4) inversion of DNA sequence that carries the promoter for one flagellin structural gene and for a repressor of a second flagellin gene • Occurs by virtue of a DNA invertase called Hin • Promotes site-specific recombination between two closely linked sites of DNA P hin Inverted repeats H 2 flagellin Repressor H 1 flagellin
- Slides: 27