GENETIC LINKAGE and CROSSING OVER Genetic linkage is




- Slides: 40

GENETIC LINKAGE and CROSSING OVER

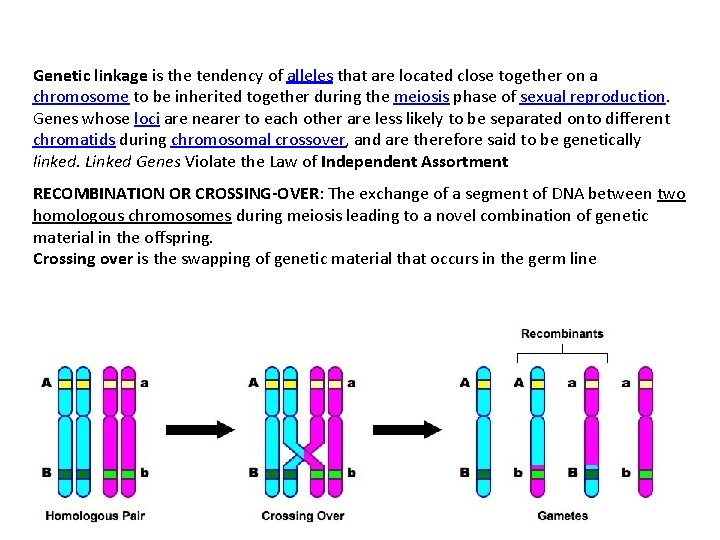
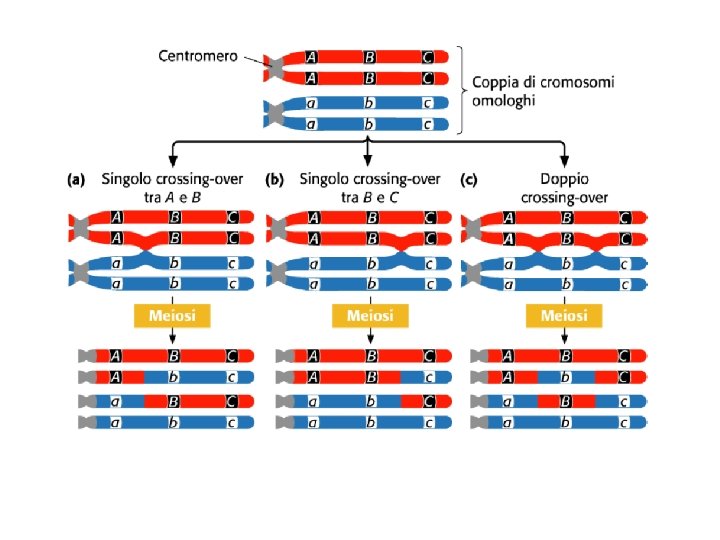
Genetic linkage is the tendency of alleles that are located close together on a chromosome to be inherited together during the meiosis phase of sexual reproduction. Genes whose loci are nearer to each other are less likely to be separated onto different chromatids during chromosomal crossover, and are therefore said to be genetically linked. Linked Genes Violate the Law of Independent Assortment RECOMBINATION OR CROSSING-OVER: The exchange of a segment of DNA between two homologous chromosomes during meiosis leading to a novel combination of genetic material in the offspring. Crossing over is the swapping of genetic material that occurs in the germ line

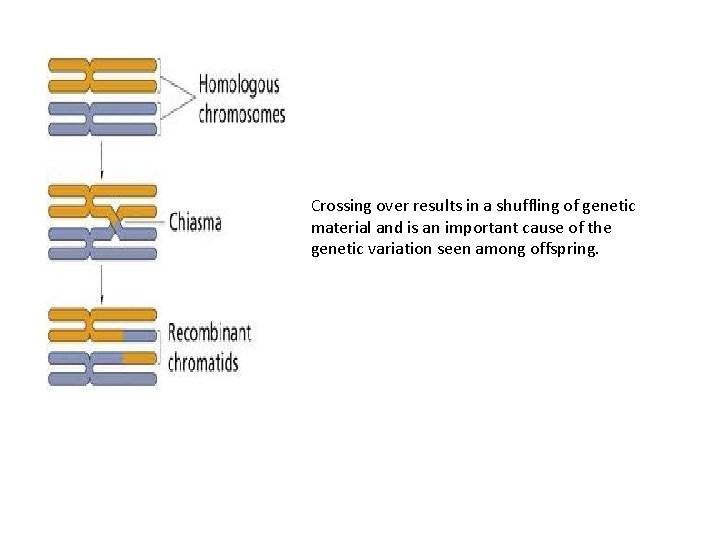
Crossing over results in a shuffling of genetic material and is an important cause of the genetic variation seen among offspring.

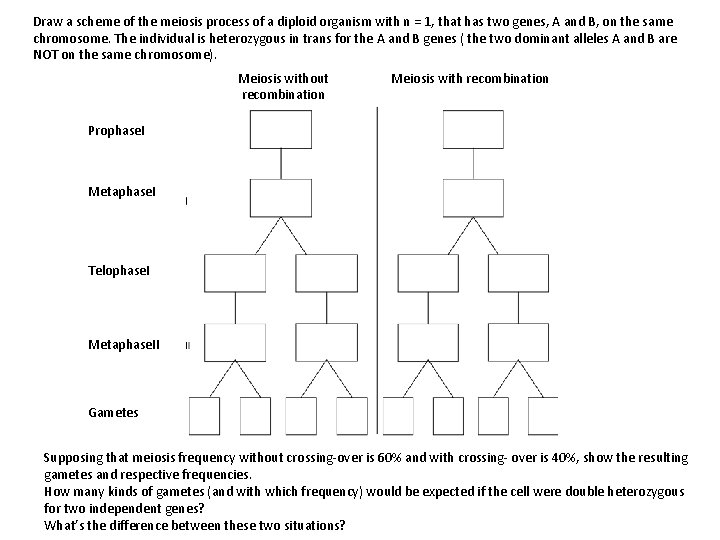
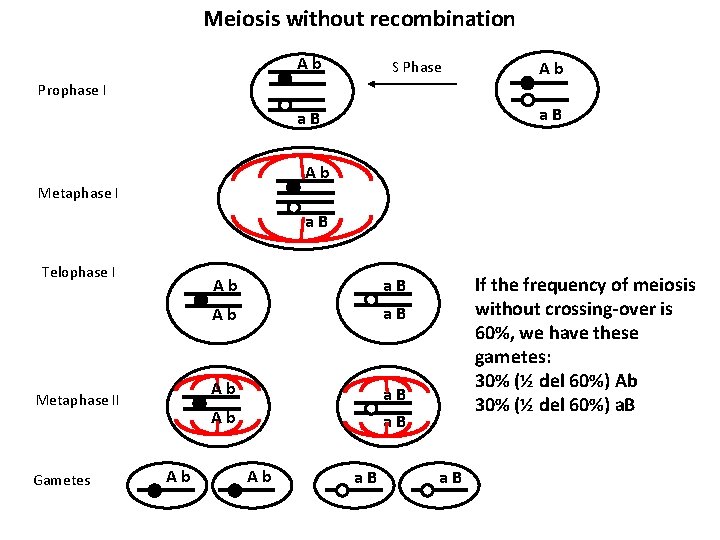
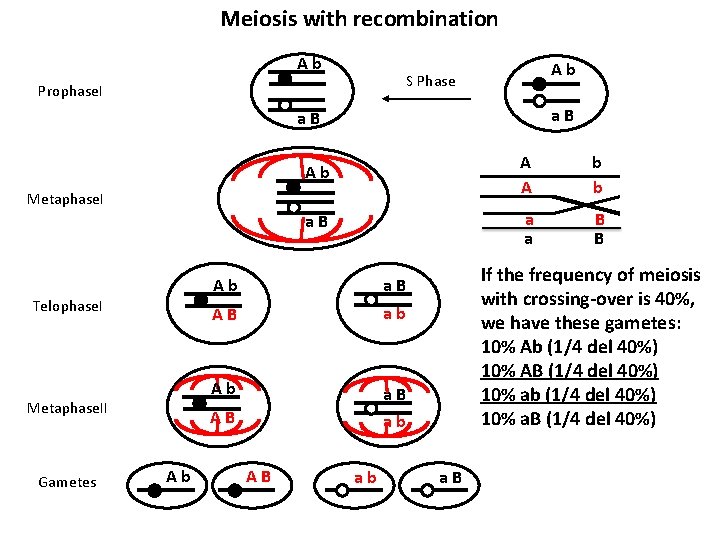
Draw a scheme of the meiosis process of a diploid organism with n = 1, that has two genes, A and B, on the same chromosome. The individual is heterozygous in trans for the A and B genes ( the two dominant alleles A and B are NOT on the same chromosome). Meiosis without recombination Meiosis with recombination Prophase. I Metaphase. I Telophase. I Metaphase. II Gametes Supposing that meiosis frequency without crossing-over is 60% and with crossing- over is 40%, show the resulting gametes and respective frequencies. How many kinds of gametes (and with which frequency) would be expected if the cell were double heterozygous for two independent genes? What’s the difference between these two situations?

Meiosis without recombination A b S Phase A b Prophase I a B A b Metaphase I a B Telophase I Metaphase II Gametes A b a B A b a B If the frequency of meiosis without crossing-over is 60%, we have these gametes: 30% (½ del 60%) Ab 30% (½ del 60%) a. B a B

Meiosis with recombination A b S Phase Prophase. I a B A b A b a B a B Metaphase. I Telophase. I Metaphase. II Gametes A b a B A B a b A B A b a b If the frequency of meiosis with crossing-over is 40%, we have these gametes: 10% Ab (1/4 del 40%) 10% AB (1/4 del 40%) 10% ab (1/4 del 40%) 10% a. B (1/4 del 40%) a B

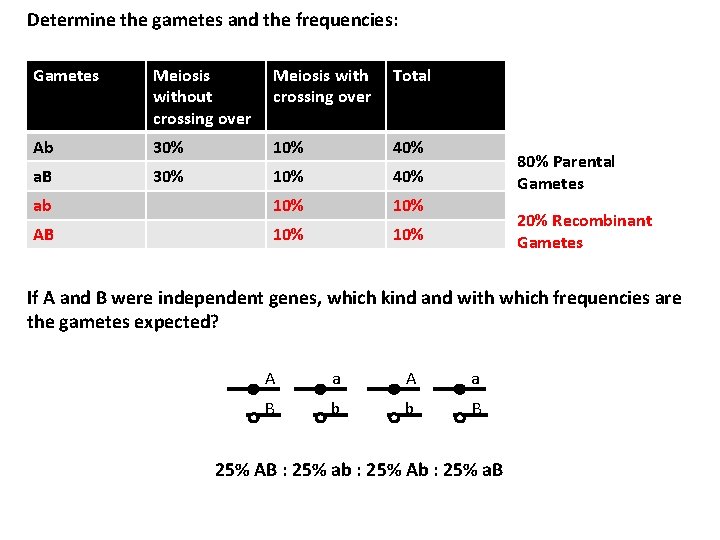
Determine the gametes and the frequencies: Gametes Meiosis without crossing over Meiosis with crossing over Total Ab 30% 10% 40% a. B 30% 10% 40% ab 10% AB 10% 80% Parental Gametes 20% Recombinant Gametes If A and B were independent genes, which kind and with which frequencies are the gametes expected? A a B b b B 25% AB : 25% ab : 25% Ab : 25% a. B

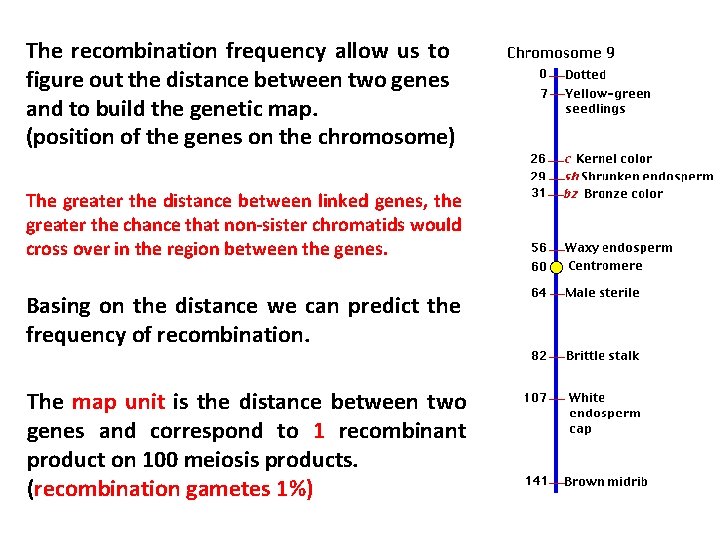
The recombination frequency allow us to figure out the distance between two genes and to build the genetic map. (position of the genes on the chromosome) The greater the distance between linked genes, the greater the chance that non-sister chromatids would cross over in the region between the genes. Basing on the distance we can predict the frequency of recombination. The map unit is the distance between two genes and correspond to 1 recombinant product on 100 meiosis products. (recombination gametes 1%)

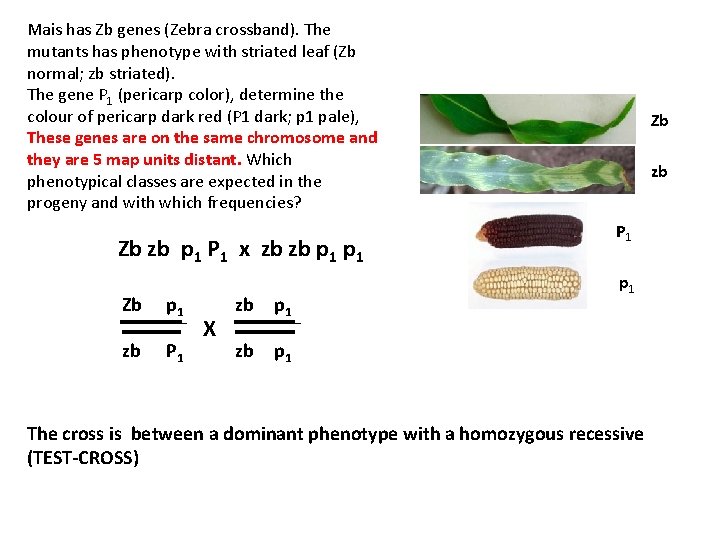
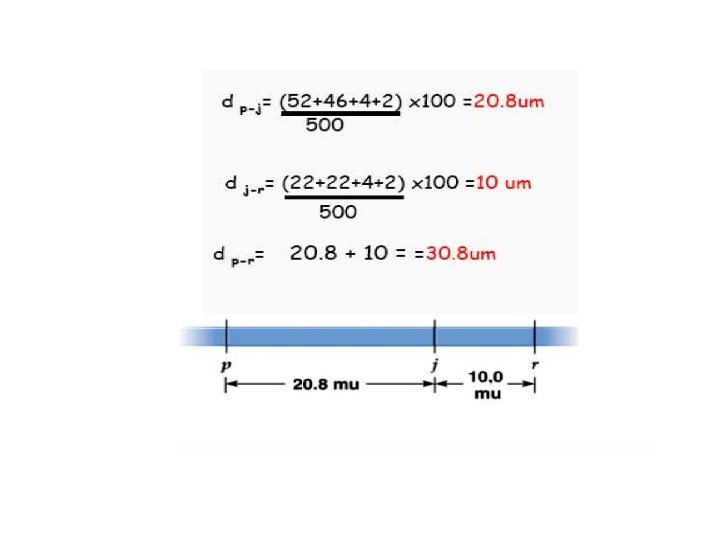
Mais has Zb genes (Zebra crossband). The mutants has phenotype with striated leaf (Zb normal; zb striated). The gene P 1 (pericarp color), determine the colour of pericarp dark red (P 1 dark; p 1 pale), These genes are on the same chromosome and they are 5 map units distant. Which phenotypical classes are expected in the progeny and with which frequencies? Zb zb p 1 P 1 x zb zb p 1 Zb p 1 zb P 1 X zb p 1 Zb zb P 1 p 1 zb p 1 The cross is between a dominant phenotype with a homozygous recessive (TEST-CROSS)

Which are the parental classes? Which are the recombinant classes? The heterozygous parent produces these gametes Zb zb X p 1 P 1 X zb p 1 Parental gametes Zb p 1 e zb P 1 95% Recombinant gametes Zb P 1 e zb p 1 5% Distance of 5 mu then frequency of recombinant gametes is 5% Each recombinant gamete is expected with a frequency of 2, 5% The parental gametes are 95: 2= 47, 5% each Parental class= during the meiosis, recombination process did not happen the gametes derived from the original structure. Recombinant class= during the meiosis recombination process happens (crossing-over)

Determine the phenotypic classes of the progeny: Zb p 1 X zb P 1 X zb p 1 The homozygous recessive produces only gametes : zb p 1 The final phenotypes are: Frequency Phenotype 47, 5% Zb zb p 1 Normal Leaf, pale pericarp 47, 5% zb zb P 1 p 1 Striated Leaf, dark pericarp 2, 5% Zb zb P 1 p 1 Normal Leaf, dark pericarp 2, 5% zb zb- p 1 Striated Leaf, pale pericarp

In drosophila gene b (black body) and gene vg (vestigial wings) are 18 mu distant. The dominant alleles are b+ (brown colour) and vg+ (normal wings), the recessive alleles b (black colour) and vg (vestigial wings). A individual b+b+ vg+vg+ is crossed with an individual bb vgvg. Draw the map P b vg b+ vg+ X 18 b+ vg+ 18 b vg b+ vg+ F 1 18 b vg F 1 will be double heterozygous with genes in cis and dominant phenotype.

If an individual of F 1 is crossed with an recessive homozygous, which phenotypic classes are expected in the progeny and with which frequencies? Gametes of the heterozygous parent are: Parental b+ vg+ e b vg Recombinant b+ vg e b vg+ b+ vg+ X b vg The frequencies depend on the distance between 2 genes: we know that the distance is 18 mu. The recombination gametes will be 18% The recombinant gametes are 18 : 2 = 9% each The parental gametes are 100 – 18 = 82 : 2 = 41% each The recessive parent produces only b vg gametes Then the expected phenotype are: Frequency Phenotype class 41% b+- vg+- Brown body, normal wings 41% b -vg- Black body, vestigial wings 9% b+ -vg- Brown body, vestigial wings 9% b- vg+- Black body, normal wings

4. If the genes A and B are located on the same chromosome at distance of 20 map units, which percentage of the cells has done crossing-over during meiosis? 1 Cell with crossing–over event = 2 rec. Gametes (1/2) and 2 parental gametes (1/2). Recombinant gametes = 20%, Cells with meiosis and crossing-over are = 20 x 2 = 40%cells c-o = 20% ric and 20% parental 60%cells no c-o = 60% parental

Parent Phenotypes AB X ab TOTAL Progeny phenotypes AB 23 Ab 21 a. B 17 ab 18 79 1) The second parent is double homozygous recessive 2) The first parent is heterozygous for both genes 3) Genotypes Aa Bb x aa bb 2) Expected phenotype classes (if the genes are independent) ¼ AB : ¼ Ab : ¼ a. B : ¼ ab Total of 79 individuals: we expected 19, 75 individuals for each class.

We can compare the expected progeny with the observed progeny and figure out the c 2 Phenotypes Xo H Xa (Xo – Xa)2 : Xa c 2 AB 23 ¼ 19, 75 10, 56 19, 75 0, 53 Ab 21 ¼ 19, 75 1, 56 19, 75 0, 08 a. B 17 ¼ 19, 75 7, 56 19, 75 0, 38 ab 18 ¼ 19, 75 3, 06 19, 75 0, 15 Totale 79 4/4 79 1, 14 GL = 4 -1 = 3 c 2 = 1, 14, PROBABILITY = 70% / 80% Conclusion: we accept the hypothesis of INDEPENDENT GENES A B a b 4) Scheme:

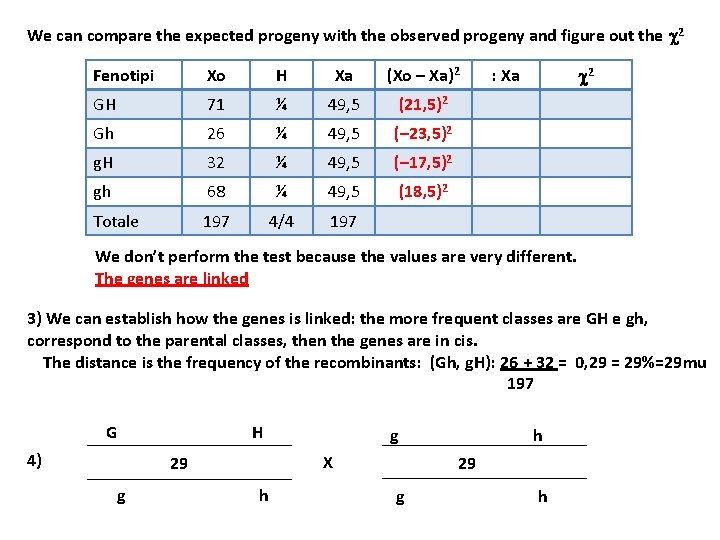
Parent phenotypes GH X gh TOTAL Phenotypes of progeny GH 71 Gh 26 g. H 32 gh 68 197 1) Second parent must be homozygous recessive for both characters. 2) First parent must be double heterozygous (in progeny we have the recessive phenotypes) 3) Genotypes Gg Hh x gg hh 4) Expected phenotypic classes ¼ GH : ¼ Gh : ¼ g. H : ¼ gh total of 197, we expect 49, 25 individual for each class

We can compare the expected progeny with the observed progeny and figure out the c 2 Fenotipi Xo H Xa (Xo – Xa)2 GH 71 ¼ 49, 5 (21, 5)2 Gh 26 ¼ 49, 5 (– 23, 5)2 g. H 32 ¼ 49, 5 (– 17, 5)2 gh 68 ¼ 49, 5 (18, 5)2 Totale 197 4/4 197 : Xa c 2 We don’t perform the test because the values are very different. The genes are linked 3) We can establish how the genes is linked: the more frequent classes are GH e gh, correspond to the parental classes, then the genes are in cis. The distance is the frequency of the recombinants: (Gh, g. H): 26 + 32 = 0, 29 = 29%=29 mu 197 G H 4) 29 g h X 29 g h

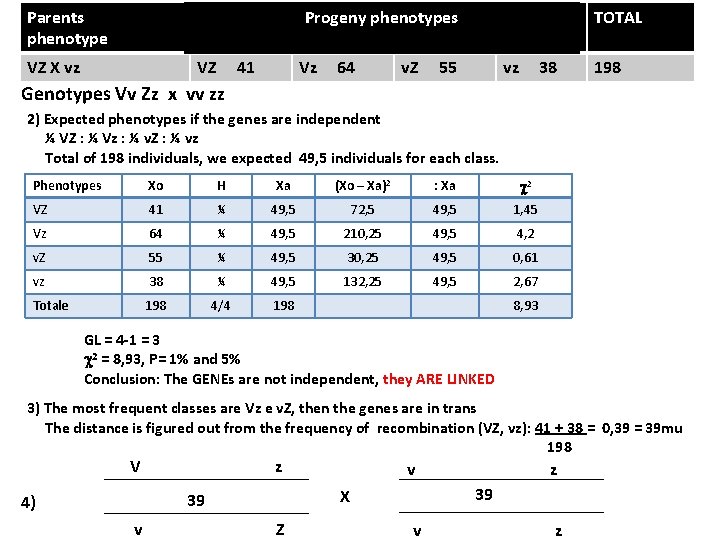
Progeny phenotypes Parents phenotype VZ X vz VZ 41 Vz 64 v. Z 55 TOTAL vz 38 198 Genotypes Vv Zz x vv zz 2) Expected phenotypes if the genes are independent ¼ VZ : ¼ Vz : ¼ v. Z : ¼ vz Total of 198 individuals, we expected 49, 5 individuals for each class. Phenotypes Xo H Xa (Xo – Xa)2 : Xa c 2 VZ 41 ¼ 49, 5 72, 5 49, 5 1, 45 Vz 64 ¼ 49, 5 210, 25 49, 5 4, 2 v. Z 55 ¼ 49, 5 30, 25 49, 5 0, 61 vz 38 ¼ 49, 5 132, 25 49, 5 2, 67 Totale 198 4/4 198 8, 93 GL = 4 -1 = 3 c 2 = 8, 93, P= 1% and 5% Conclusion: The GENEs are not independent, they ARE LINKED 3) The most frequent classes are Vz e v. Z, then the genes are in trans The distance is figured out from the frequency of recombination (VZ, vz): 41 + 38 = 0, 39 = 39 mu 198 V z 4) 39 v Z X v z 39 v z

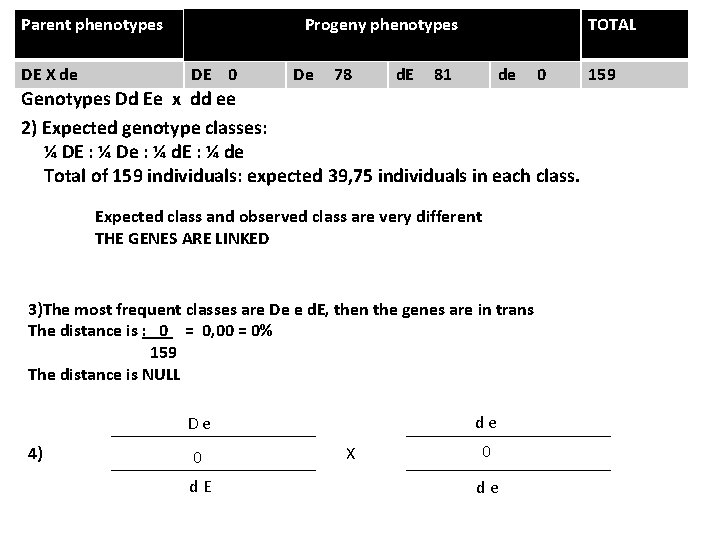
Parent phenotypes DE X de TOTAL Progeny phenotypes DE 0 De 78 d. E 81 de 0 Genotypes Dd Ee x dd ee 2) Expected genotype classes: ¼ DE : ¼ De : ¼ d. E : ¼ de Total of 159 individuals: expected 39, 75 individuals in each class. Expected class and observed class are very different THE GENES ARE LINKED 3)The most frequent classes are De e d. E, then the genes are in trans The distance is : 0 = 0, 00 = 0% 159 The distance is NULL d e D e 4) 0 d E X 0 d e 159

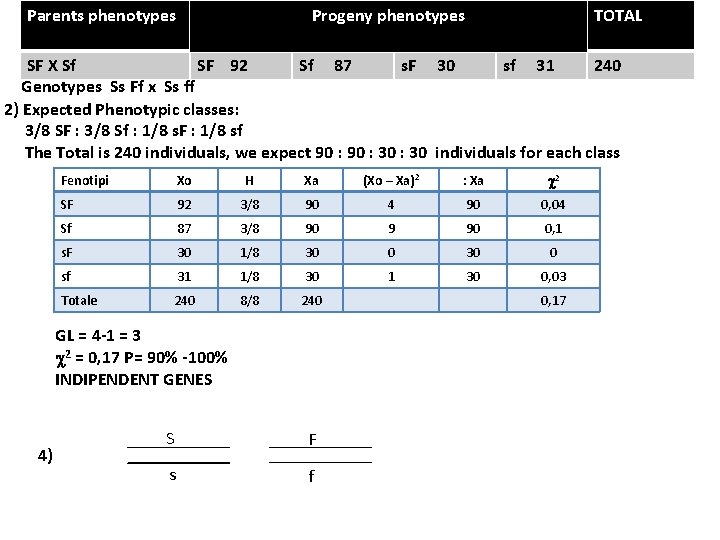
Parents phenotypes TOTAL Progeny phenotypes SF X Sf SF 92 Sf 87 s. F 30 sf 31 240 Genotypes Ss Ff x Ss ff 2) Expected Phenotypic classes: 3/8 SF : 3/8 Sf : 1/8 s. F : 1/8 sf The Total is 240 individuals, we expect 90 : 30 individuals for each class Fenotipi Xo H Xa (Xo – Xa)2 : Xa c 2 SF 92 3/8 90 4 90 0, 04 Sf 87 3/8 90 9 90 0, 1 s. F 30 1/8 30 0 sf 31 1/8 30 1 30 0, 03 Totale 240 8/8 240 GL = 4 -1 = 3 c 2 = 0, 17 P= 90% -100% INDIPENDENT GENES 4) S F s f 0, 17

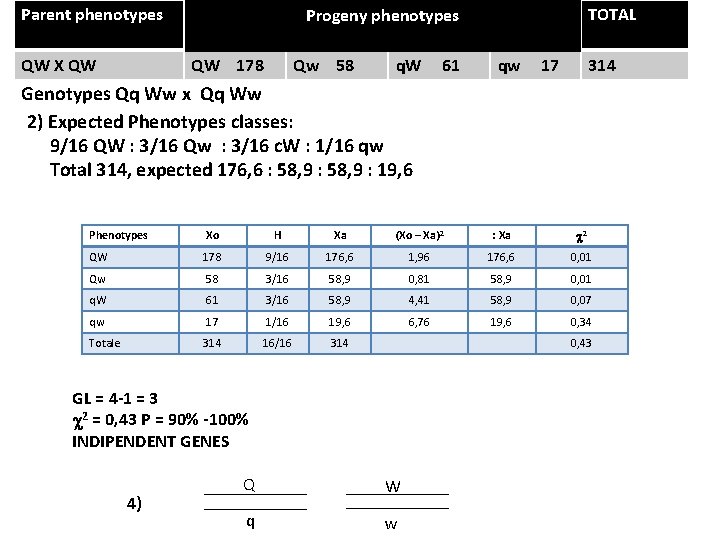
Parent phenotypes QW X QW TOTAL Progeny phenotypes QW 178 Qw 58 q. W 61 qw 17 314 Genotypes Qq Ww x Qq Ww 2) Expected Phenotypes classes: 9/16 QW : 3/16 Qw : 3/16 c. W : 1/16 qw Total 314, expected 176, 6 : 58, 9 : 19, 6 Phenotypes Xo H Xa (Xo – Xa)2 : Xa c 2 QW 178 9/16 176, 6 1, 96 176, 6 0, 01 Qw 58 3/16 58, 9 0, 81 58, 9 0, 01 q. W 61 3/16 58, 9 4, 41 58, 9 0, 07 qw 17 1/16 19, 6 6, 76 19, 6 0, 34 Totale 314 16/16 314 0, 43 GL = 4 -1 = 3 c 2 = 0, 43 P = 90% -100% INDIPENDENT GENES 4) Q W q w

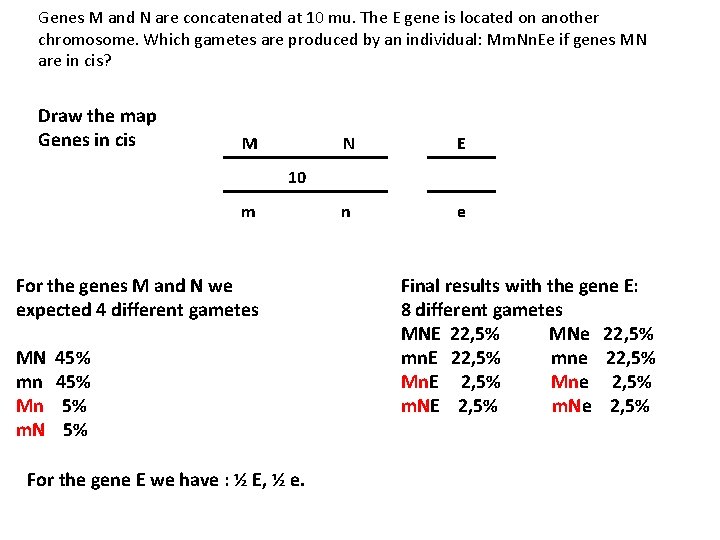
Genes M and N are concatenated at 10 mu. The E gene is located on another chromosome. Which gametes are produced by an individual: Mm. Nn. Ee if genes MN are in cis? Draw the map Genes in cis M N E 10 m n For the genes M and N we expected 4 different gametes MN 45% mn 45% Mn 5% m. N 5% For the gene E we have : ½ E, ½ e. e Final results with the gene E: 8 different gametes MNE 22, 5% MNe 22, 5% mn. E 22, 5% mne 22, 5% Mn. E 2, 5% Mne 2, 5% m. NE 2, 5% m. Ne 2, 5%

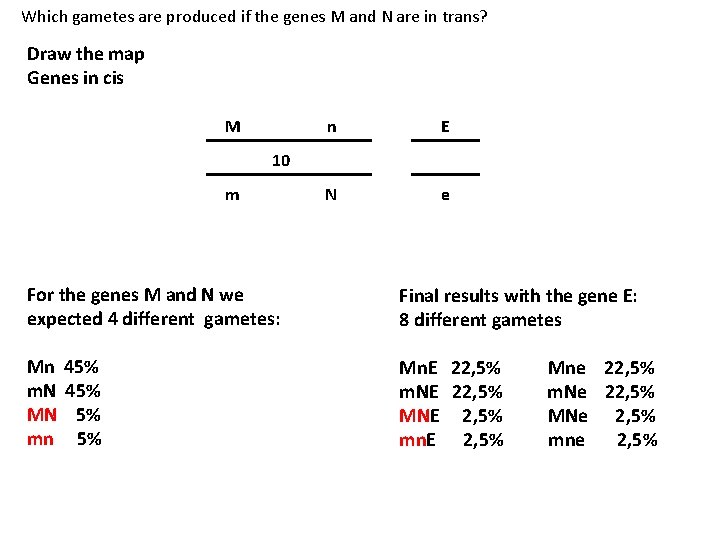
Which gametes are produced if the genes M and N are in trans? Draw the map Genes in cis M n E 10 m N e For the genes M and N we expected 4 different gametes: Final results with the gene E: 8 different gametes Mn 45% m. N 45% MN 5% mn 5% Mn. E 22, 5% Mne 22, 5% m. NE 22, 5% m. Ne 22, 5% MNE 2, 5% MNe 2, 5% mn. E 2, 5% mne 2, 5%

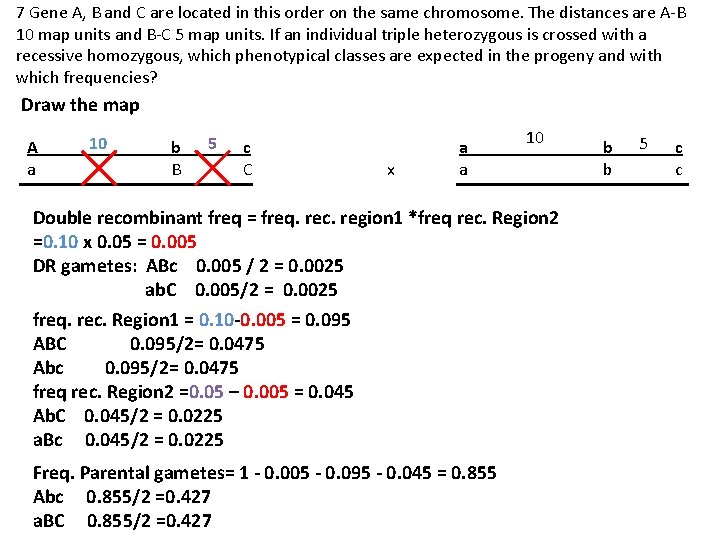
7 Gene A, B and C are located in this order on the same chromosome. The distances are A-B 10 map units and B-C 5 map units. If an individual triple heterozygous is crossed with a recessive homozygous, which phenotypical classes are expected in the progeny and with which frequencies? Draw the map A a 10 b B 5 c C x a a 10 Double recombinant freq = freq. rec. region 1 *freq rec. Region 2 =0. 10 x 0. 05 = 0. 005 DR gametes: ABc 0. 005 / 2 = 0. 0025 ab. C 0. 005/2 = 0. 0025 freq. rec. Region 1 = 0. 10 -0. 005 = 0. 095 ABC 0. 095/2= 0. 0475 Abc 0. 095/2= 0. 0475 freq rec. Region 2 =0. 05 – 0. 005 = 0. 045 Ab. C 0. 045/2 = 0. 0225 a. Bc 0. 045/2 = 0. 0225 Freq. Parental gametes= 1 - 0. 005 - 0. 095 - 0. 045 = 0. 855 Abc 0. 855/2 =0. 427 a. BC 0. 855/2 =0. 427 b b 5 c c

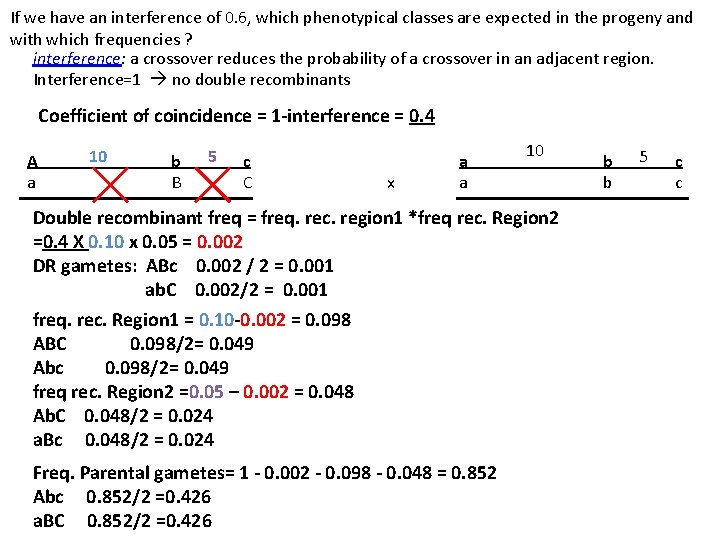
If we have an interference of 0. 6, which phenotypical classes are expected in the progeny and with which frequencies ? interference: a crossover reduces the probability of a crossover in an adjacent region. Interference=1 no double recombinants Coefficient of coincidence = 1 -interference = 0. 4 A a 10 b B 5 c C x a a 10 Double recombinant freq = freq. rec. region 1 *freq rec. Region 2 =0. 4 X 0. 10 x 0. 05 = 0. 002 DR gametes: ABc 0. 002 / 2 = 0. 001 ab. C 0. 002/2 = 0. 001 freq. rec. Region 1 = 0. 10 -0. 002 = 0. 098 ABC 0. 098/2= 0. 049 Abc 0. 098/2= 0. 049 freq rec. Region 2 =0. 05 – 0. 002 = 0. 048 Ab. C 0. 048/2 = 0. 024 a. Bc 0. 048/2 = 0. 024 Freq. Parental gametes= 1 - 0. 002 - 0. 098 - 0. 048 = 0. 852 Abc 0. 852/2 =0. 426 a. BC 0. 852/2 =0. 426 b b 5 c c

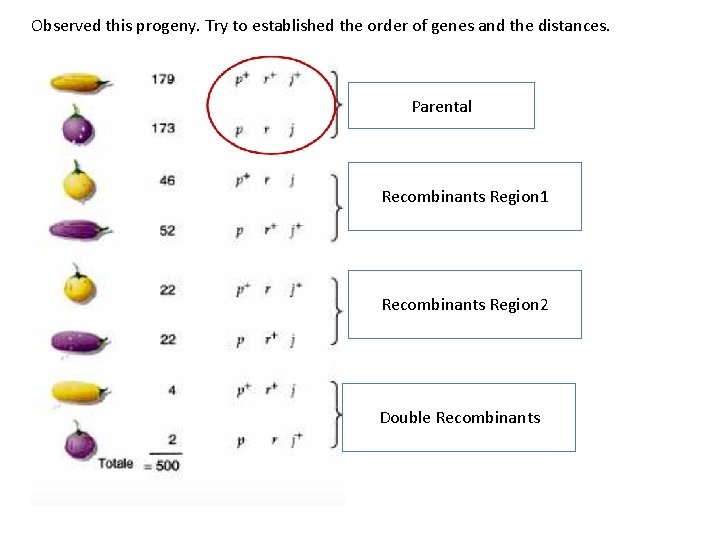
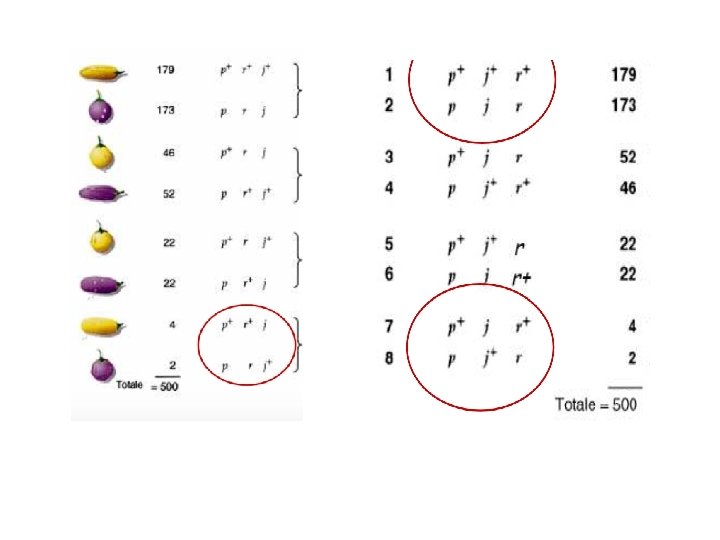
Observed this progeny. Try to established the order of genes and the distances. Parental Recombinants Region 1 Recombinants Region 2 Double Recombinants




GENE INTERACTIONS

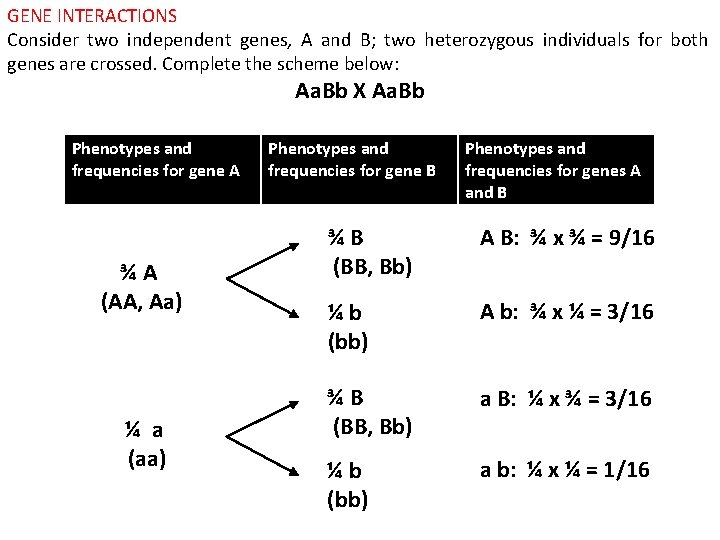
GENE INTERACTIONS Consider two independent genes, A and B; two heterozygous individuals for both genes are crossed. Complete the scheme below: Aa. Bb X Aa. Bb Phenotypes and frequencies for gene A ¾ A (AA, Aa) ¼ a (aa) Phenotypes and frequencies for gene B Phenotypes and frequencies for genes A and B ¾ B (BB, Bb) A B: ¾ x ¾ = 9/16 ¼ b (bb) A b: ¾ x ¼ = 3/16 ¾ B (BB, Bb) a B: ¼ x ¾ = 3/16 ¼ b (bb) a b: ¼ x ¼ = 1/16

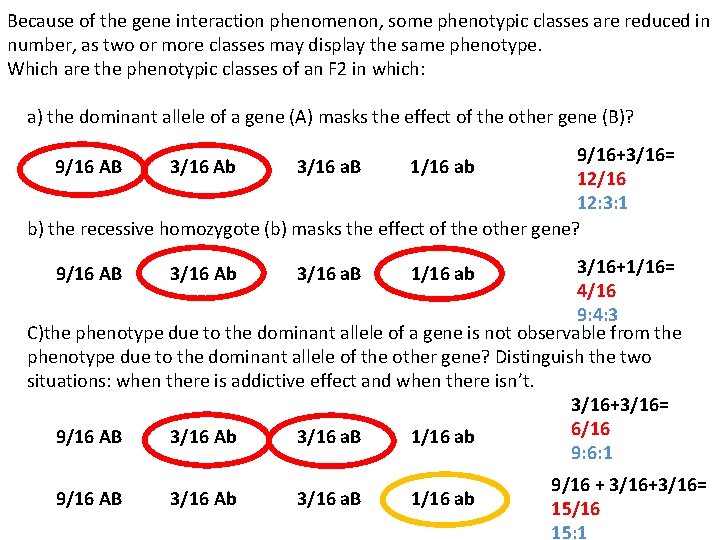
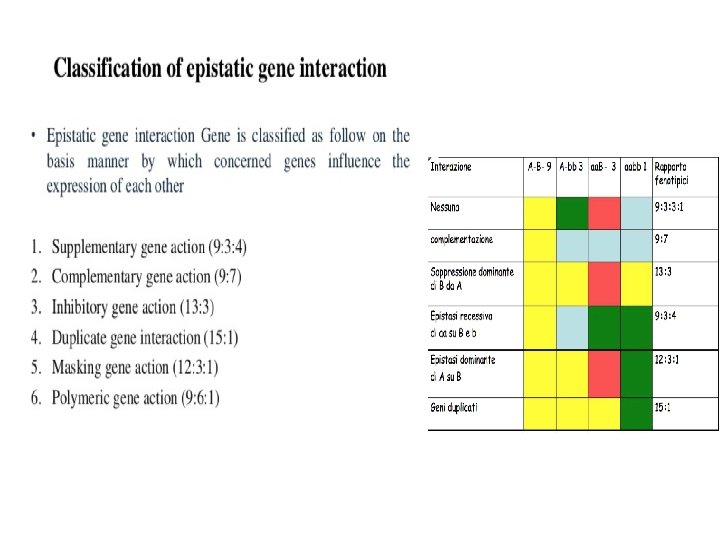
Because of the gene interaction phenomenon, some phenotypic classes are reduced in number, as two or more classes may display the same phenotype. Which are the phenotypic classes of an F 2 in which: a) the dominant allele of a gene (A) masks the effect of the other gene (B)? 9/16+3/16= 12/16 12: 3: 1 b) the recessive homozygote (b) masks the effect of the other gene? 9/16 AB 3/16 Ab 3/16 a. B 1/16 ab 3/16+1/16= 4/16 9: 4: 3 C)the phenotype due to the dominant allele of a gene is not observable from the phenotype due to the dominant allele of the other gene? Distinguish the two situations: when there is addictive effect and when there isn’t. 3/16+3/16= 6/16 9/16 AB 3/16 Ab 3/16 a. B 1/16 ab 9: 6: 1 9/16 AB 3/16 Ab 3/16 a. B 1/16 ab 9/16 + 3/16+3/16= 15/16 15: 1

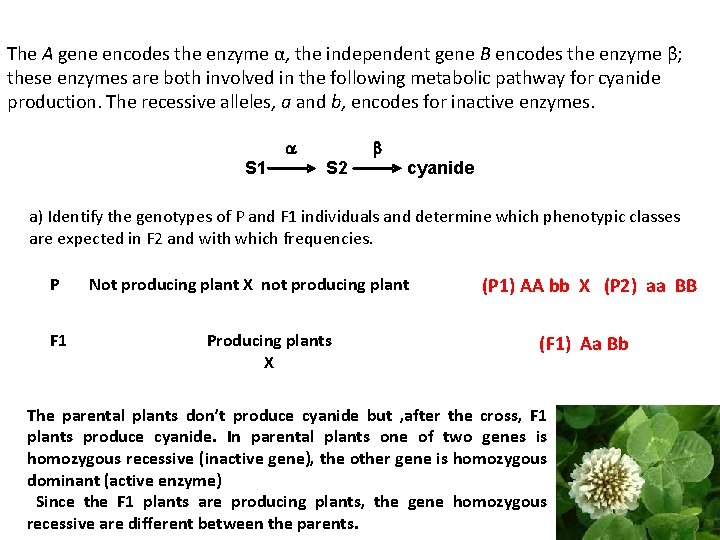
The A gene encodes the enzyme α, the independent gene B encodes the enzyme β; these enzymes are both involved in the following metabolic pathway for cyanide production. The recessive alleles, a and b, encodes for inactive enzymes. a S 1 b S 2 cyanide a) Identify the genotypes of P and F 1 individuals and determine which phenotypic classes are expected in F 2 and with which frequencies. P F 1 Not producing plant X not producing plant Producing plants X (P 1) AA bb X (P 2) aa BB (F 1) Aa Bb The parental plants don’t produce cyanide but , after the cross, F 1 plants produce cyanide. In parental plants one of two genes is homozygous recessive (inactive gene), the other gene is homozygous dominant (active enzyme) Since the F 1 plants are producing plants, the gene homozygous recessive are different between the parents.

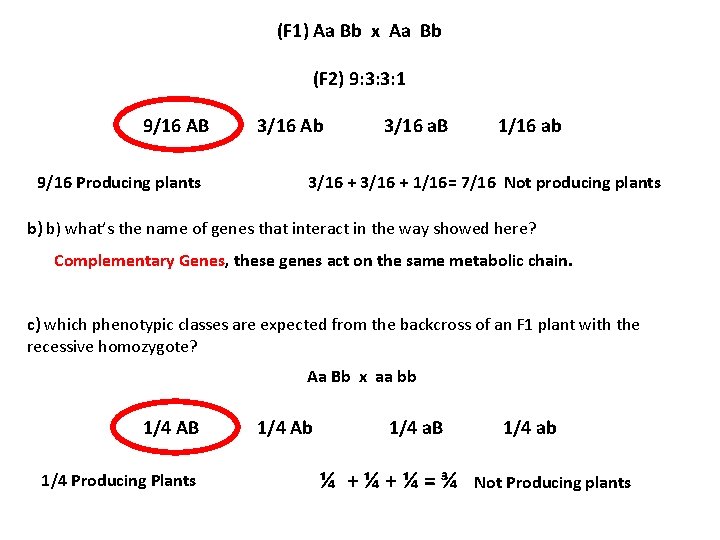
(F 1) Aa Bb x Aa Bb (F 2) 9: 3: 3: 1 9/16 AB 9/16 Producing plants 3/16 Ab 3/16 a. B 1/16 ab 3/16 + 1/16= 7/16 Not producing plants b) b) what’s the name of genes that interact in the way showed here? Complementary Genes, these genes act on the same metabolic chain. c) which phenotypic classes are expected from the backcross of an F 1 plant with the recessive homozygote? Aa Bb x aa bb 1/4 AB 1/4 Producing Plants 1/4 Ab 1/4 a. B 1/4 ab ¼ + ¼ = ¾ Not Producing plants

The two independent genes P and C control two different consecutive steps in the same metabolic pathway. P C S 1 S 2 Yellow fruit The dominant alleles P and C encode for active enzymes, while the recessive alleles, p and c, encode for inactive enzymes. Which phenotype is expected for individuals with the following genotype (intermediate substrates are colourless): a) PPcc This plant produces the enzyme encoded by gene P, but not produces the other one (gene C). Metabolic chain is blocked at step S 2. Phenotype: white melon b) Pp. Cc The plant has both enzymes thus the metabolic chain works until the end. Phenotype: yellow melon. c) pp. CC The plant hasn’t the enzyme encoded by gene P. It produces the enzyme encoded by the gene C but it doesn’t convert S 1 in S 2. Phenotype white melon. d) Ppcc The plant produces the first enzyme but not the second one. Then the metabolic chain is blocked at S 1 step. Phenotype white melon.

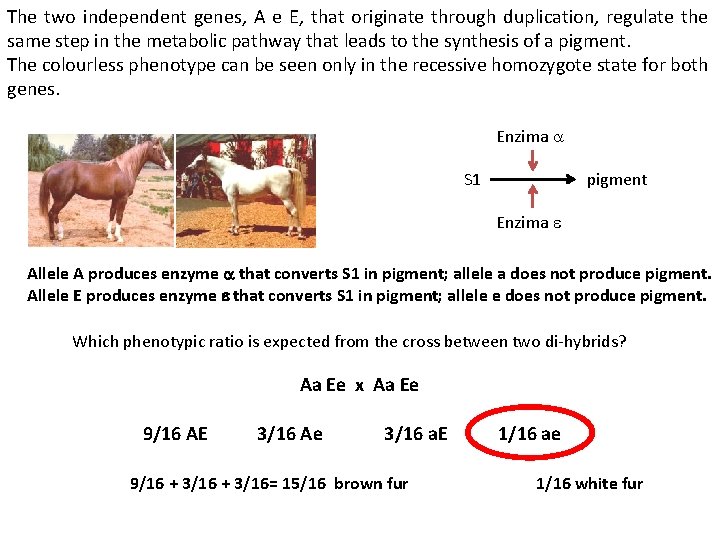
The two independent genes, A e E, that originate through duplication, regulate the same step in the metabolic pathway that leads to the synthesis of a pigment. The colourless phenotype can be seen only in the recessive homozygote state for both genes. Enzima a S 1 pigment Enzima e Allele A produces enzyme a that converts S 1 in pigment; allele a does not produce pigment. Allele E produces enzyme e that converts S 1 in pigment; allele e does not produce pigment. Which phenotypic ratio is expected from the cross between two di-hybrids? Aa Ee x Aa Ee 9/16 AE 3/16 Ae 3/16 a. E 9/16 + 3/16= 15/16 brown fur 1/16 ae 1/16 white fur

Two pure lines of pepper are crossed: first line with Red fruits and the second one with orange fruits. The plants of F 1 produce only red peppers, but crossing two individuals of F 1 we obtain: 192 RED FRUITS, 47 ORANGE FRUITS, 14 WHITE FRUITS Pure lines = they pass the same alleles to all offsprings and so they are homozygous We know that the colour fruit is controlled by 2 genes (A and B), which kind of interaction is present? Determine the genotypes and hypothesize a possible action on metabolic chain. (P 1) RED FRUITS X (P 2) ORANGE FRUITS AA bb aa BB (F 1) RED FRUITS Aa Bb The dominant allele A is epistatic on gene B F 2: 9/16 AB + 3/16 Ab 192 red fruits 47 orange fruits 3/16 a. B 14 white fruits 1/16 ab When the plant has an allele of A the phenotype related to gene B is not shown. Phenotypes AB and Ab produce red fruits When A is homozygous recessive, the plant show the B phenotype: if it has dominant allele B the fruits will be orange, if it is homozygous bb the fruits will be white. 192+47+14= 253 (: 16= 15. 8)

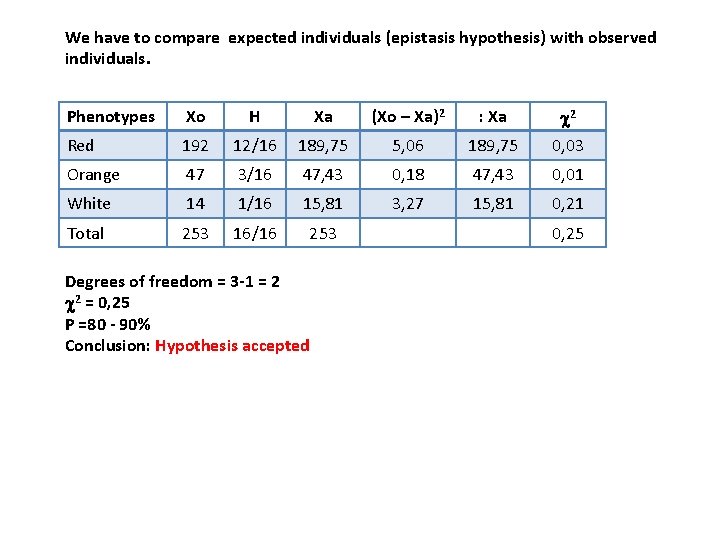
We have to compare expected individuals (epistasis hypothesis) with observed individuals. Phenotypes Xo H Xa (Xo – Xa)2 : Xa c 2 Red 192 12/16 189, 75 5, 06 189, 75 0, 03 Orange 47 3/16 47, 43 0, 18 47, 43 0, 01 White 14 1/16 15, 81 3, 27 15, 81 0, 21 Total 253 16/16 253 Degrees of freedom = 3 -1 = 2 c 2 = 0, 25 P =80 - 90% Conclusion: Hypothesis accepted 0, 25
