Gene Structure and Identification Eukaryotic Genes and Genomes













- Slides: 32

Gene Structure and Identification • Eukaryotic Genes and Genomes • Gene Finding Previous reading: 1. 3, 9. 1 -9. 6 Reading: 10. 2, 10. 4, 10. 6 -8 BIO 520 Bioinformatics Jim Lund

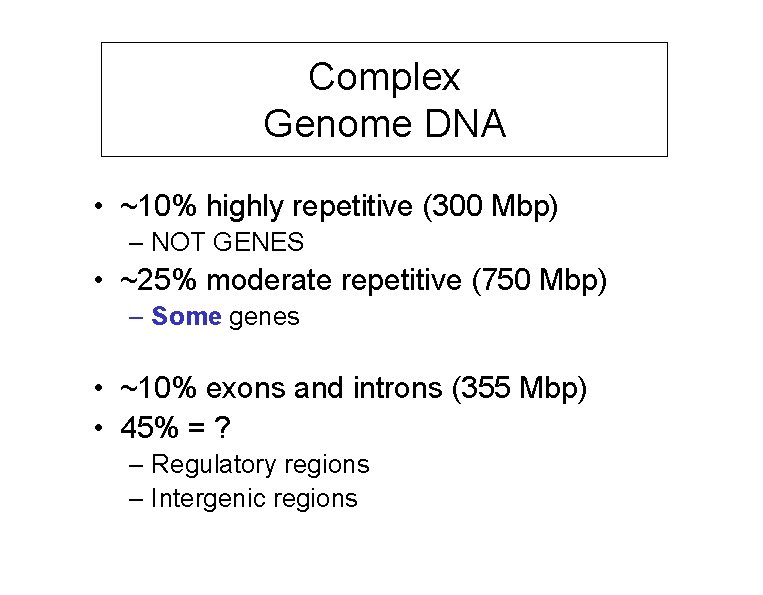
Complex Genome DNA • ~10% highly repetitive (300 Mbp) – NOT GENES • ~25% moderate repetitive (750 Mbp) – Some genes • ~10% exons and introns (355 Mbp) • 45% = ? – Regulatory regions – Intergenic regions

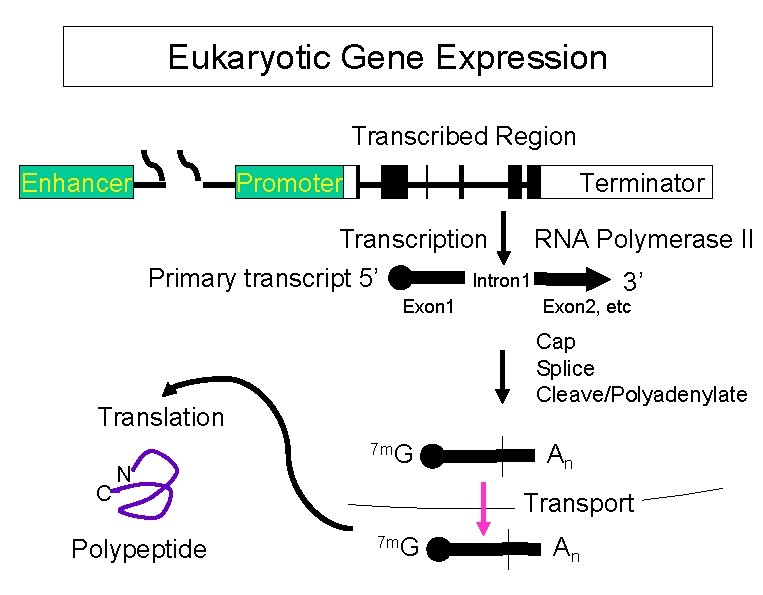
Eukaryotic Gene Expression Transcribed Region Enhancer Promoter Terminator Transcription Primary transcript 5’ Polypeptide Exon 2, etc Cap Splice Cleave/Polyadenylate Translation C 3’ Intron 1 Exon 1 N RNA Polymerase II 7 m. G An Transport 7 m. G An

Yeast • ORFS = genes! Small ORFS (RNA genes) Regulatory Sequences

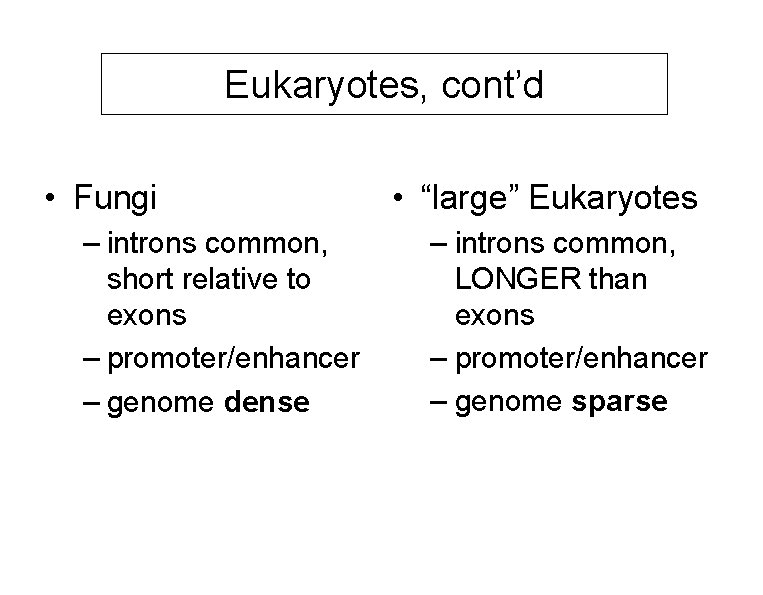
Eukaryotes, cont’d • Fungi – introns common, short relative to exons – promoter/enhancer – genome dense • “large” Eukaryotes – introns common, LONGER than exons – promoter/enhancer – genome sparse

% of genes Intron Prevalence Introns

% of genes Intron Size Introns

% of genes Exon Size Exon size (bps)

Fungi Sew together exons –ORF regions –consensus sequences –domain/polypeptide matches

Exon/Intron Structure CCACATTgtn(30 -10, 000)an(5 -20)ag. CAGAA. . . CCACATTCAGAA. . . Pro. His. Ser. Glu. . .

Alternative Splice CCACATTgtn(30 -10, 000)an(5 -20)agcag. AA . . . CCACATTAA. . . Pro. His. STOP

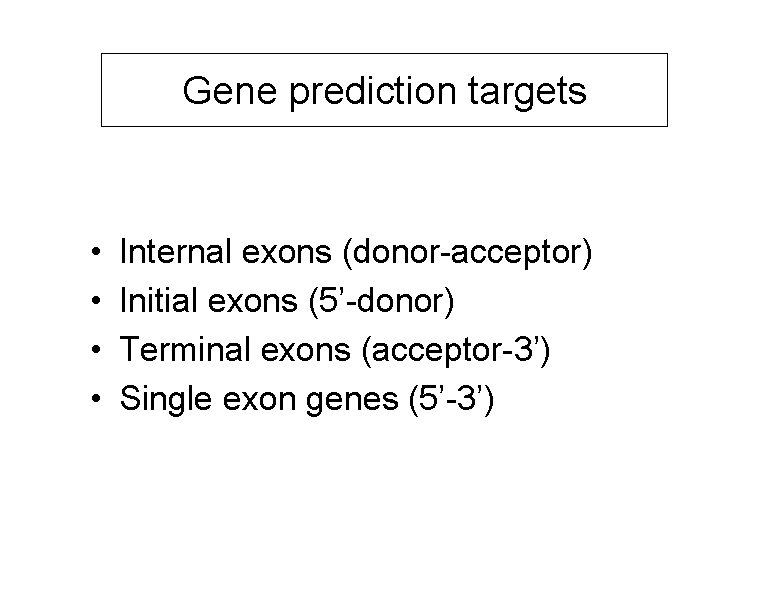
Gene prediction targets • • Internal exons (donor-acceptor) Initial exons (5’-donor) Terminal exons (acceptor-3’) Single exon genes (5’-3’)

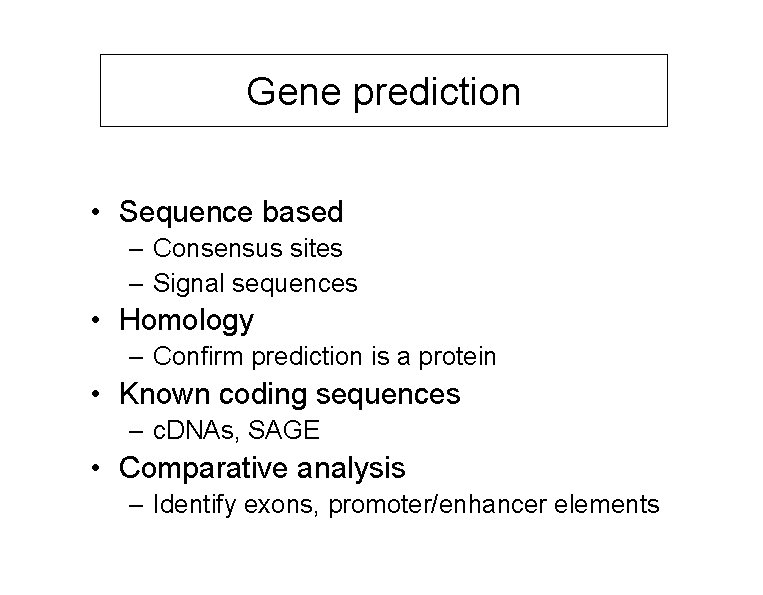
Gene prediction • Sequence based – Consensus sites – Signal sequences • Homology – Confirm prediction is a protein • Known coding sequences – c. DNAs, SAGE • Comparative analysis – Identify exons, promoter/enhancer elements

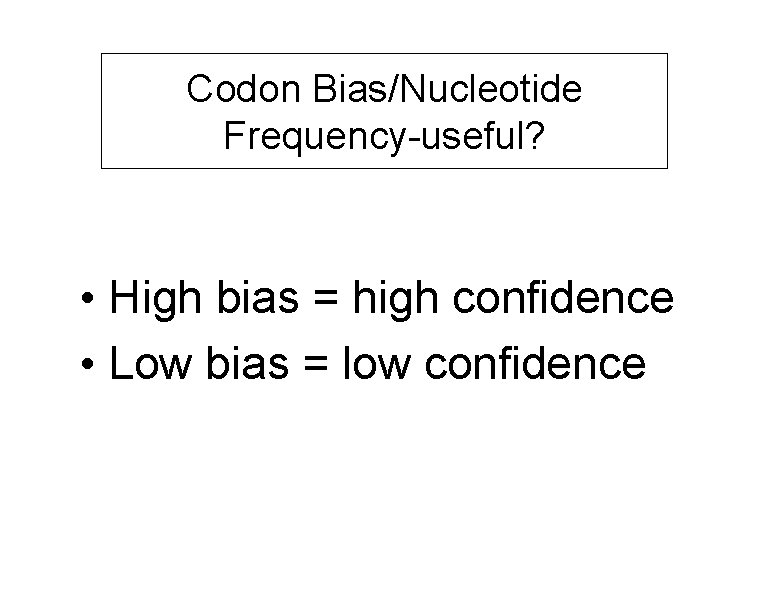
Codon Bias/Nucleotide Frequency-useful? • High bias = high confidence • Low bias = low confidence

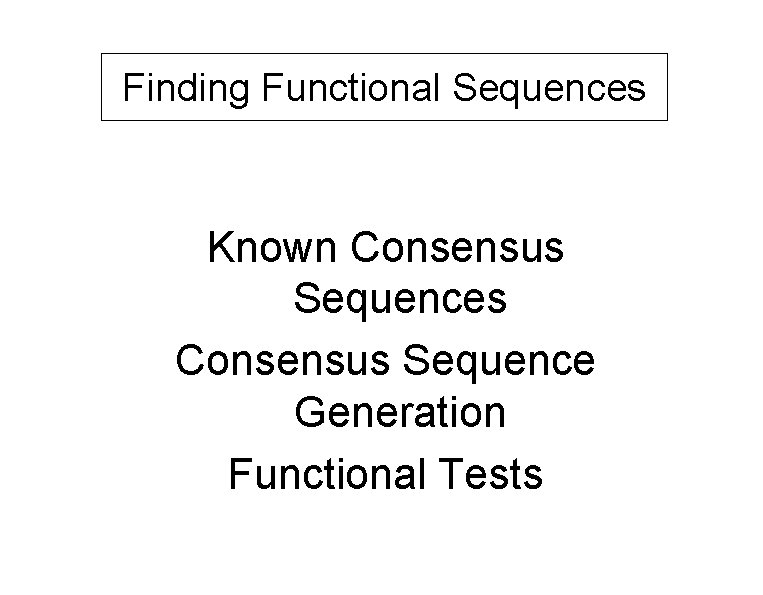
Finding Functional Sequences Known Consensus Sequences Consensus Sequence Generation Functional Tests

Describing consensus sequences • Position Weight Matrices – Sequence Logos • Hidden Markov Models

Translation Initiation Sites

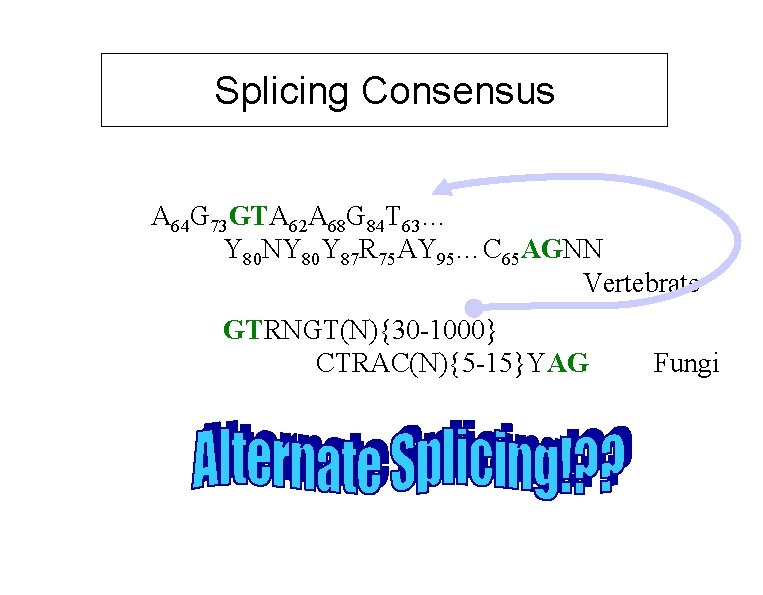
Splicing Consensus A 64 G 73 GTA 62 A 68 G 84 T 63… Y 80 NY 80 Y 87 R 75 AY 95…C 65 AGNN Vertebrate GTRNGT(N){30 -1000} CTRAC(N){5 -15}YAG Fungi

Linguistic approach to combining gene features • Non-repetitive DNA!! • Long ORF – similar to known protein • ORF extended by “reasonable” splices • ORF begins with “good” ATG • Promoter/terminator flanks

DATABASE SEARCH • BLASTN – DNA: DNA comparison (ALWAYS!) – Not sensitive (DNA conservation low) • BLASTX/TBLASTX – 6 frame ORFS: polypeptide database – 6 frames vs. 6 frames of a DNA database

Protein Database Matches Very helpful for the “known” What about the unknown? ? ?

Transcript Initiation • Basal Promoters • Enhancers/Silencers/Regulatory Sites – Boundary elements? • Transcription Initation Prokaryotes vs Eukaryotes Organism-to-Organism

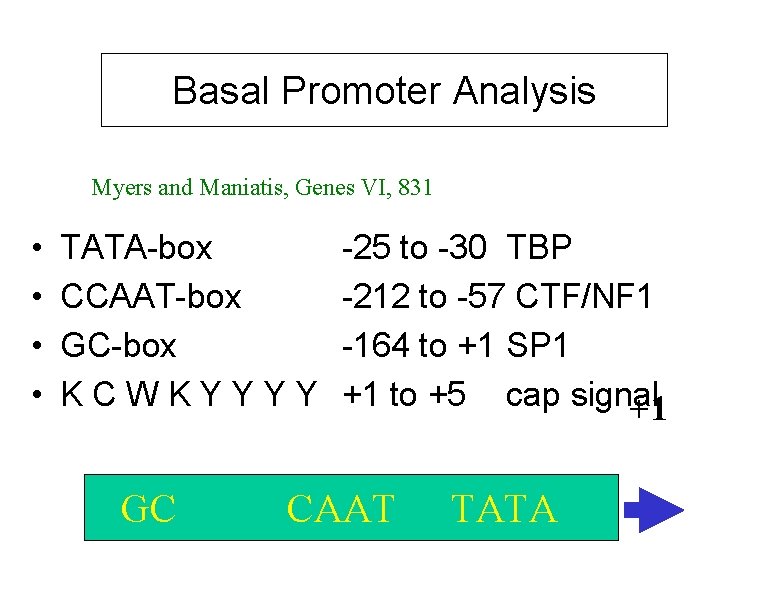
Basal Promoter Analysis Myers and Maniatis, Genes VI, 831 • • TATA-box CCAAT-box GC-box KCWKYYYY GC -25 to -30 TBP -212 to -57 CTF/NF 1 -164 to +1 SP 1 +1 to +5 cap signal +1 CAAT TATA

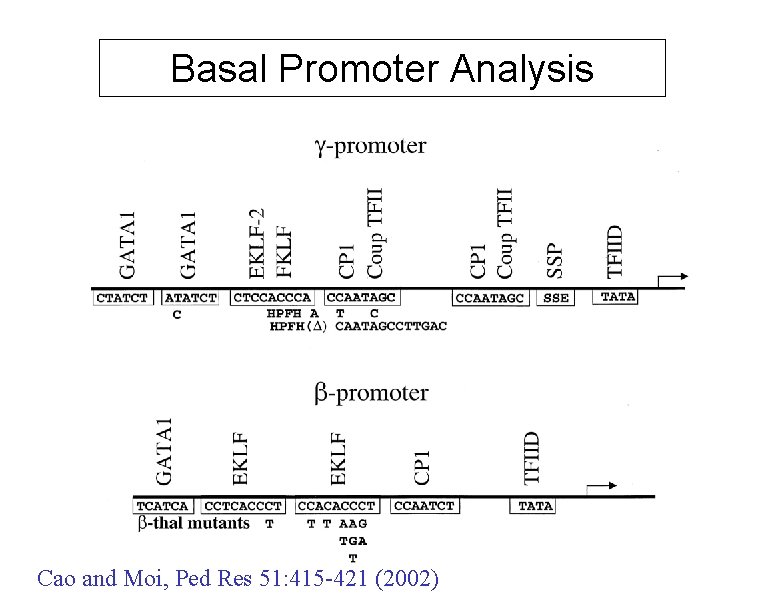
Basal Promoter Analysis Cao and Moi, Ped Res 51: 415 -421 (2002)

m. RNA processing • Exon/Intron – Alternate splicing • Polyadenylation/Cleavage • Stability

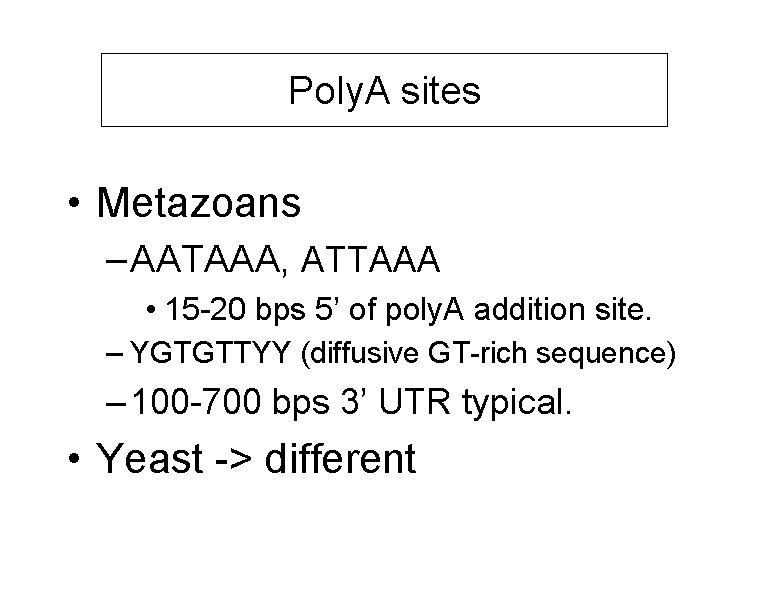
Poly. A sites • Metazoans – AATAAA, ATTAAA • 15 -20 bps 5’ of poly. A addition site. – YGTGTTYY (diffusive GT-rich sequence) – 100 -700 bps 3’ UTR typical. • Yeast -> different

Translation • Initiation site – 1 st AUG used 95% of the time. • Translational regulatory elements – translational enhancers – upstream ORFs

Tools-WWW • • • Genscan Genie GRAIL II: integrated gene parsing Gen. Lang HMMGene (lock ESTs, etc. ) GENEMARK

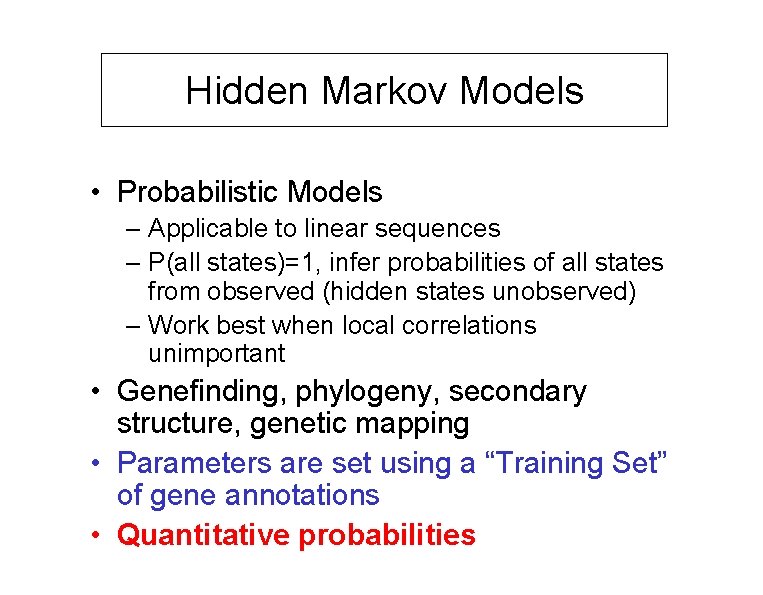
Hidden Markov Models • Probabilistic Models – Applicable to linear sequences – P(all states)=1, infer probabilities of all states from observed (hidden states unobserved) – Work best when local correlations unimportant • Genefinding, phylogeny, secondary structure, genetic mapping • Parameters are set using a “Training Set” of gene annotations • Quantitative probabilities

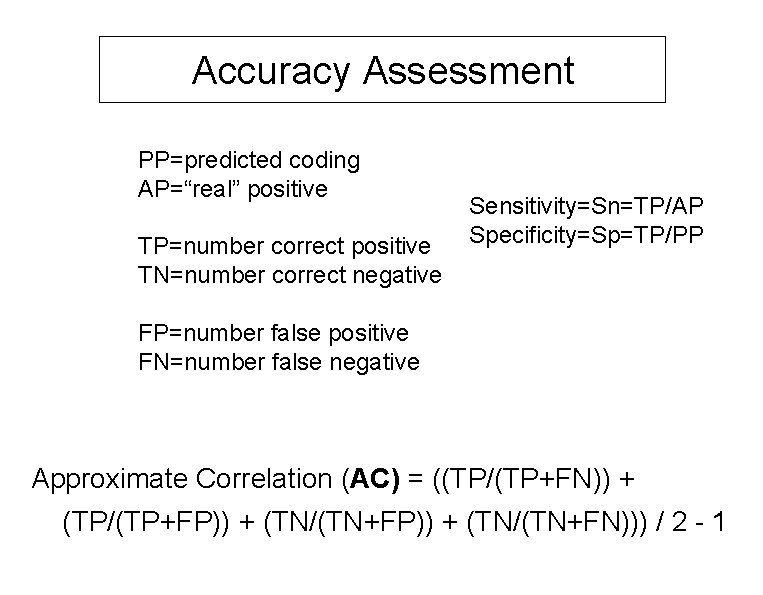
Accuracy Assessment PP=predicted coding AP=“real” positive TP=number correct positive TN=number correct negative Sensitivity=Sn=TP/AP Specificity=Sp=TP/PP FP=number false positive FN=number false negative Approximate Correlation (AC) = ((TP/(TP+FN)) + (TP/(TP+FP)) + (TN/(TN+FN))) / 2 - 1

Accuracy Levels Bp Exon

NEXT • Regulatory Sequences – Known Consensus Sequences – Consensus Sequence Generation – Functional (Lab) Data • A few examples