Gene Structure and Identification Eukaryotic Genes and Genomes





















- Slides: 45

Gene Structure and Identification Eukaryotic Genes and Genomes Gene Finding 9/17/2020 Chuck Staben 1

Complex Genome DNA • ~10% highly repetitive ( Mbp) – NOT GENES • ~25% moderate repetitive ( Mbp) – Some genes • ~25% exons and introns ( • 40%=? Mbp) – Regulatory regions – Intergenic regions 9/17/2020 Chuck Staben 2

Eukaryotic Gene Expression Engraved on your Brain!!!! 9/17/2020 Chuck Staben 3

Yeast ORFS=genes! What don’t you find this way? 9/17/2020 Chuck Staben 4

Eukaryotes, cont’d • Fungi • “large” Eukaryotes – introns – promoter/enhancer • Where? – genome dense or sparse? – Intron average= – Exon average= – Promoter/enhancer • Where/how arranged – genome sparse 9/17/2020 Chuck Staben 5

Intron Prevalence 9/17/2020 Chuck Staben 6

Intron Size 9/17/2020 Chuck Staben 7

Exon Size 9/17/2020 Chuck Staben 8

Fungi Sew together exons –ORF regions –consensus sequences –domain/polypeptide matches 9/17/2020 Chuck Staben 9

Exon/Intron Structure CCACATTgtn(30 -10, 000)an(5 -20)ag. CAGAA …________. . . Pro. His. Ser. Glu. . . 9/17/2020 Chuck Staben 10

Alternative Splice CCACATTgtn(30 -10, 000)an(5 -20)agcag. AA . . . CCACATTAA. . . Pro. His_____ 9/17/2020 Chuck Staben 11

Codon Bias/Nucleotide Frequency-useful? • Bias=0. 97 means______ • Bias=0. 03 means______ 9/17/2020 Chuck Staben 12

Consensus Sequences • Promoter sites • Intron/Exon • Transcription Termination/Poly. A • Translation initation 9/17/2020 Chuck Staben 13

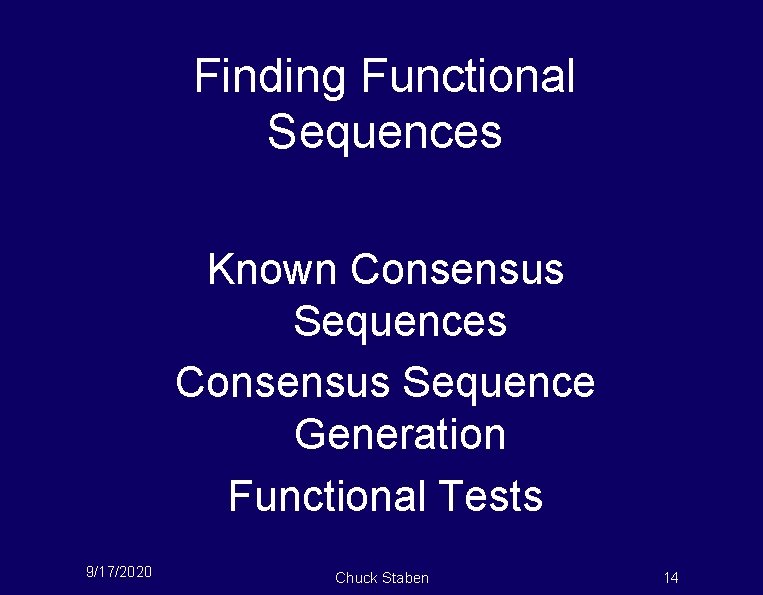
Finding Functional Sequences Known Consensus Sequences Consensus Sequence Generation Functional Tests 9/17/2020 Chuck Staben 14

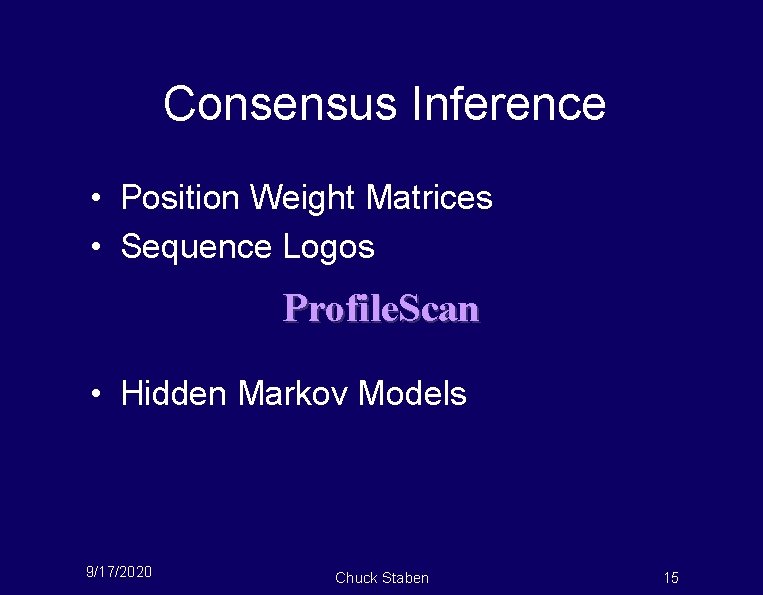
Consensus Inference • Position Weight Matrices • Sequence Logos Profile. Scan • Hidden Markov Models 9/17/2020 Chuck Staben 15

Translation Initiation Sites 9/17/2020 Chuck Staben 16

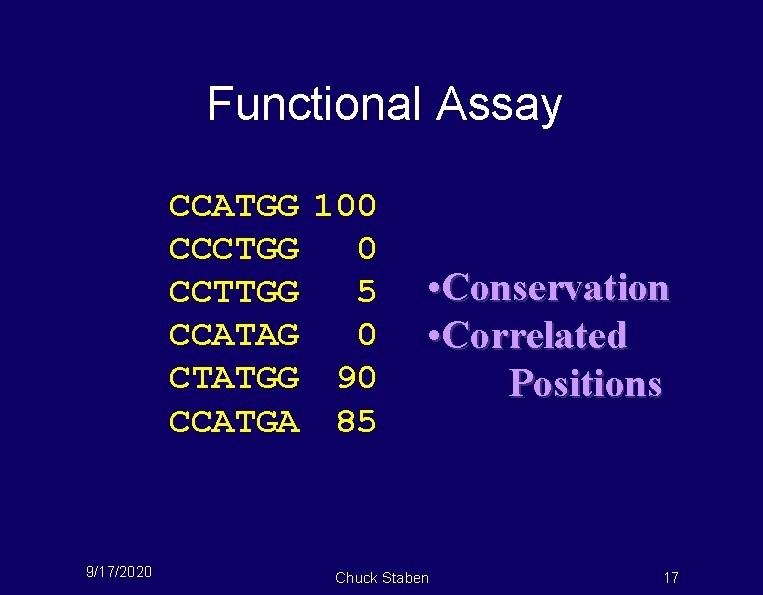
Functional Assay CCATGG 100 CCCTGG 0 CCTTGG 5 CCATAG 0 CTATGG 90 CCATGA 85 9/17/2020 • Conservation • Correlated Positions Chuck Staben 17

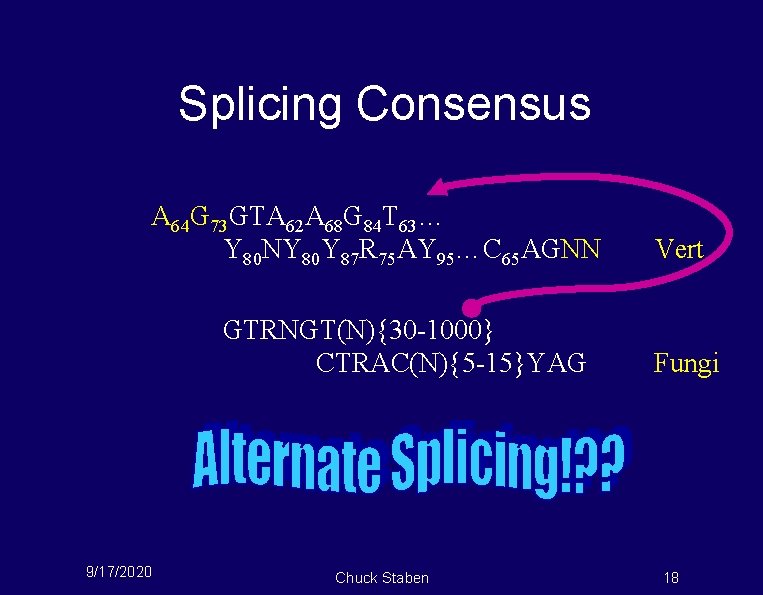
Splicing Consensus A 64 G 73 GTA 62 A 68 G 84 T 63… Y 80 NY 80 Y 87 R 75 AY 95…C 65 AGNN GTRNGT(N){30 -1000} CTRAC(N){5 -15}YAG 9/17/2020 Chuck Staben Vert Fungi 18

Linguistic Approach • Non-repetitive DNA!! • Long ORF – similar to known protein • ORF extended by “reasonable” splices • ORF begins with “good” ATG • Promoter/terminator flanks 9/17/2020 Chuck Staben 19

DATABASE SEARCH • BLASTN – What? – Limitations? • BLASTX/TBLASTX – BLASTX does? – TBALSTX? 9/17/2020 Chuck Staben 20

Protein Database Matches Great for the “known” What about the unknown? ? ? 9/17/2020 Chuck Staben 21

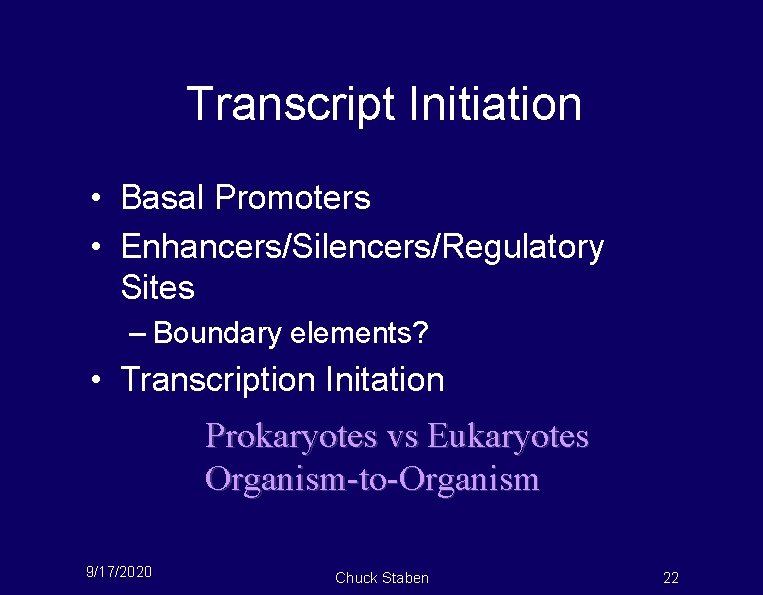
Transcript Initiation • Basal Promoters • Enhancers/Silencers/Regulatory Sites – Boundary elements? • Transcription Initation Prokaryotes vs Eukaryotes Organism-to-Organism 9/17/2020 Chuck Staben 22

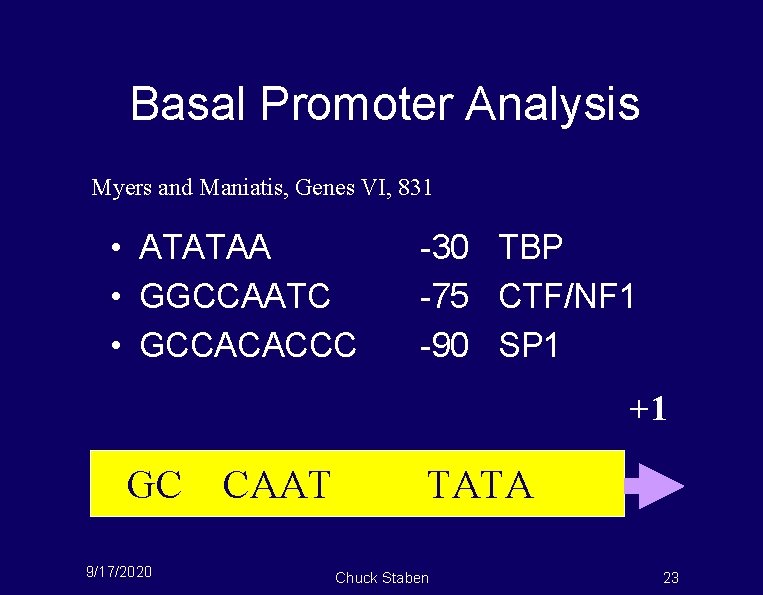
Basal Promoter Analysis Myers and Maniatis, Genes VI, 831 • ATATAA • GGCCAATC • GCCACACCC -30 TBP -75 CTF/NF 1 -90 SP 1 +1 GC 9/17/2020 CAAT TATA Chuck Staben 23

m. RNA processing • Exon/Intron –Alternate splicing • Polyadenylation/Cleavage • Stability 9/17/2020 Chuck Staben 24

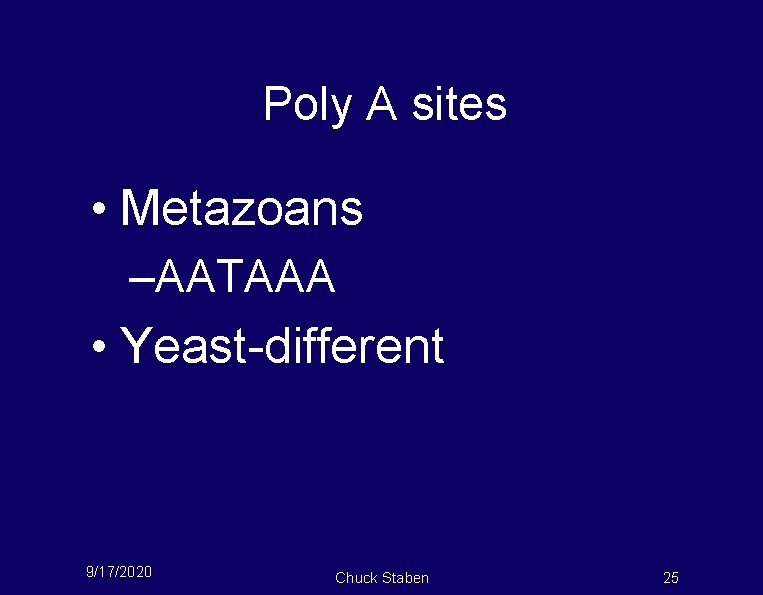
Poly A sites • Metazoans –AATAAA • Yeast-different 9/17/2020 Chuck Staben 25

Translation • Initation site • (Frameshifting) • Translational regulatory elements – upstream ORFs – translational enhancers 9/17/2020 Chuck Staben 26

Translation Sites • Initiate at 5’-ATG –upstream ORF…regulatory • (Frameshifting) • Translation enhancers…. 9/17/2020 Chuck Staben 27

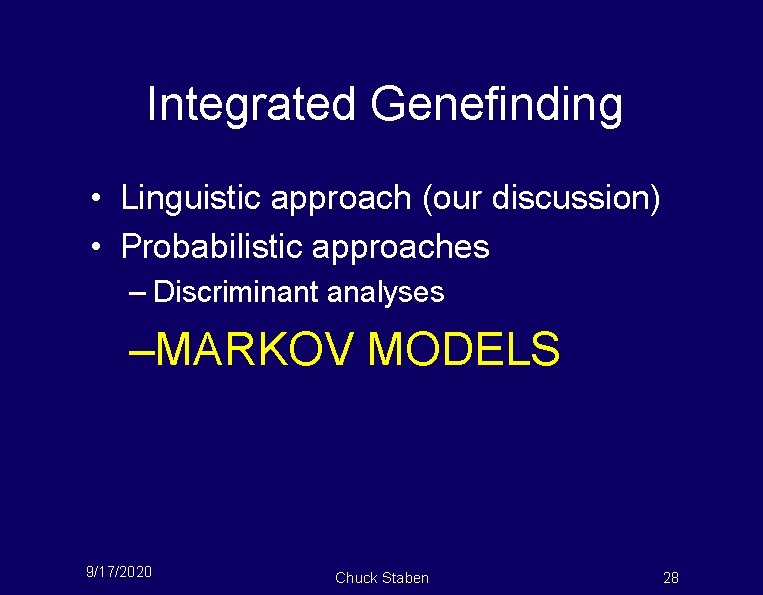
Integrated Genefinding • Linguistic approach (our discussion) • Probabilistic approaches – Discriminant analyses –MARKOV MODELS 9/17/2020 Chuck Staben 28

Tools-WWW • GRAIL II: integrated gene parsing • Gen. Lang • • GENIE HMMGene (lock ESTs, etc. ) GENSCAN GENEMARK 9/17/2020 Chuck Staben 29

Hidden Markov Models • Probabilistic Models – Applicable to linear sequences – P(all states)=1, infer probabilities of all states from observed (hidden states unobserved) – Work best when local correlations unimportant • Genefinding, phylogeny, secondary structure, genetic mapping • Work best with “Training Set” • Quantitative probabilities 9/17/2020 Chuck Staben 30

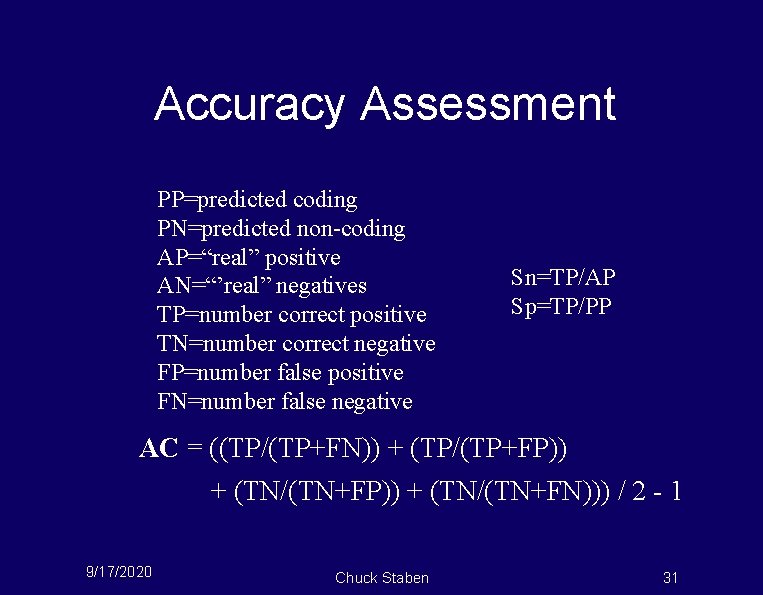
Accuracy Assessment PP=predicted coding PN=predicted non-coding AP=“real” positive AN=“’real” negatives TP=number correct positive TN=number correct negative FP=number false positive FN=number false negative Sn=TP/AP Sp=TP/PP AC = ((TP/(TP+FN)) + (TP/(TP+FP)) + (TN/(TN+FN))) / 2 - 1 9/17/2020 Chuck Staben 31

Accuracy Levels DNA Sequence Error Rate!? ? 9/17/2020 Chuck Staben 32

NEXT • Regulatory Sequences – Known Consensus Sequences – Consensus Sequence Generation – Functional (Lab) Data • Real examples 9/17/2020 Chuck Staben 33

Gene Regulatory Sequences • Functional sites –Consensus –Experimental tests • Inferred sites –Transcriptome analysis 9/17/2020 Chuck Staben 34

Regulatory Sites • Transcript initiation • m. RNA processing • Translation sites 9/17/2020 Chuck Staben 35

Regulatory Factors • lac. I, trp. R, CAP, ara. C…. • GAL 4, NDT 80… 9/17/2020 Chuck Staben 36

EUKARYOTES • • More complex signals More genes More dispersed signals Combinatoric regulation common 9/17/2020 Chuck Staben 37

Enhancer Elements • Octamer • Name some… • • • 9/17/2020 OCT 1, OCT 2 Chuck Staben 38

Consensus Sequence Databases • WWW-based –TFD (transcription factor database) –BCM Search launcher 9/17/2020 Chuck Staben 39

Transcriptome Analyses • Microarray transcription analysis • MEME analysis of clusters 9/17/2020 Chuck Staben 40

Practical Gene Finding • Use ALL tools – Comparative • BLASTN, BLASTX – Predictive: Stitch together a consensus • HMM, GRAIL… • ORF finders • Findpatterns (and WWW pattern searches) • c. DNA OR protein OR genetic evidence 9/17/2020 Chuck Staben 41

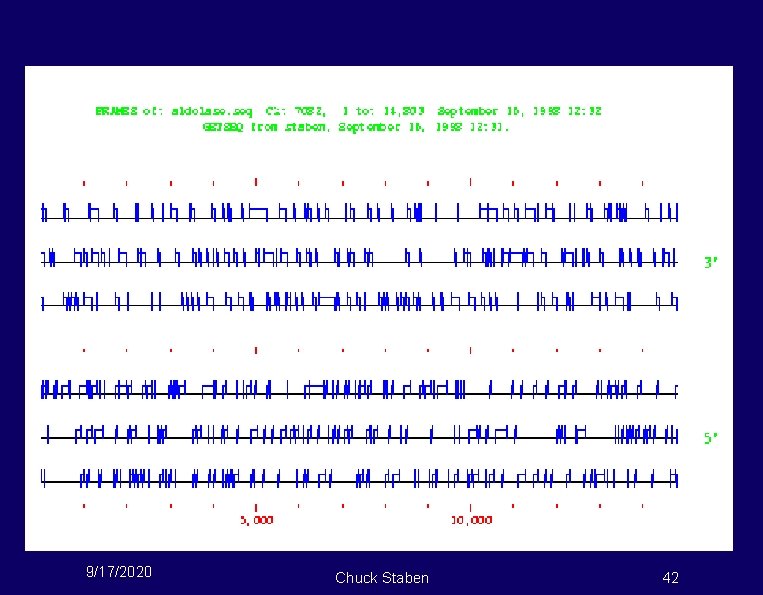
FRAMES-aldolase gene 9/17/2020 Chuck Staben 42

If aldolase is so tough, how do you really do it? Combine DNA sequence with other data! 9/17/2020 Chuck Staben 43

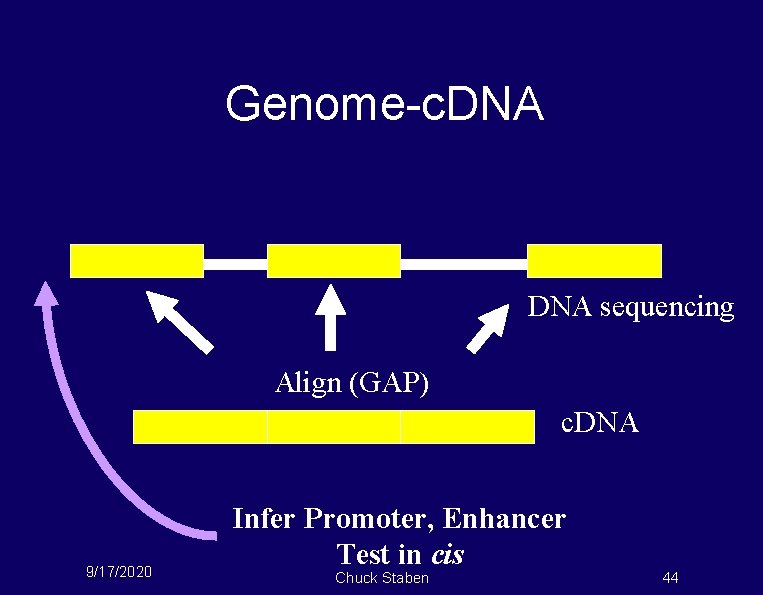
Genome-c. DNA P DNA sequencing Align (GAP) c. DNA 9/17/2020 Infer Promoter, Enhancer Test in cis Chuck Staben 44

Comparative Genomics • Conservation of coding regions • Identification of transcription signals – “words” in common 9/17/2020 Chuck Staben 45