Gene Structure 1 Topics to be Covered Introduction
Gene Structure 1

Topics to be Covered • Introduction • Gene structure – Promoter – Terminator – Splice site 2
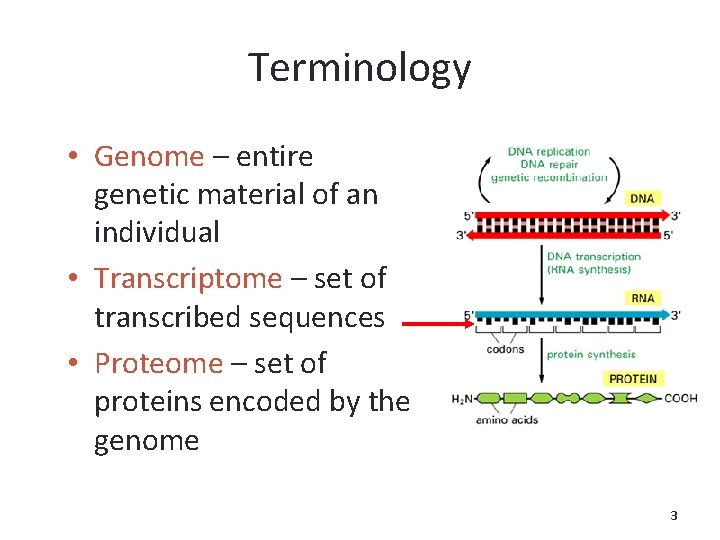
Terminology • Genome – entire genetic material of an individual • Transcriptome – set of transcribed sequences • Proteome – set of proteins encoded by the genome 3
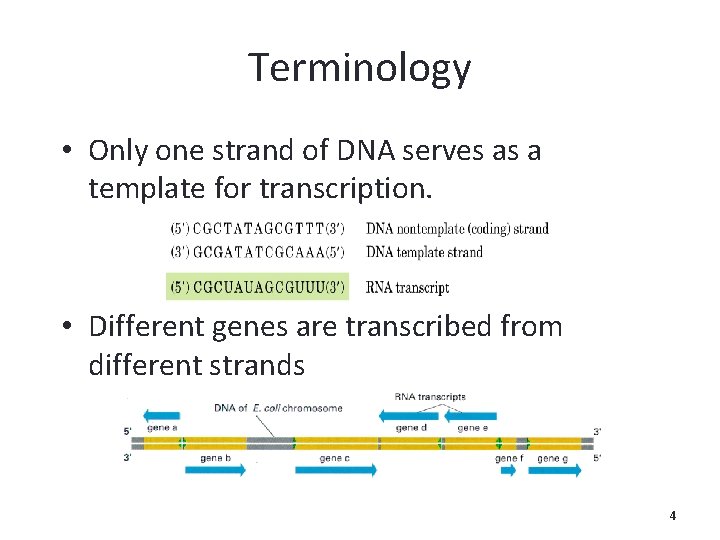
Terminology • Only one strand of DNA serves as a template for transcription. • Different genes are transcribed from different strands 4
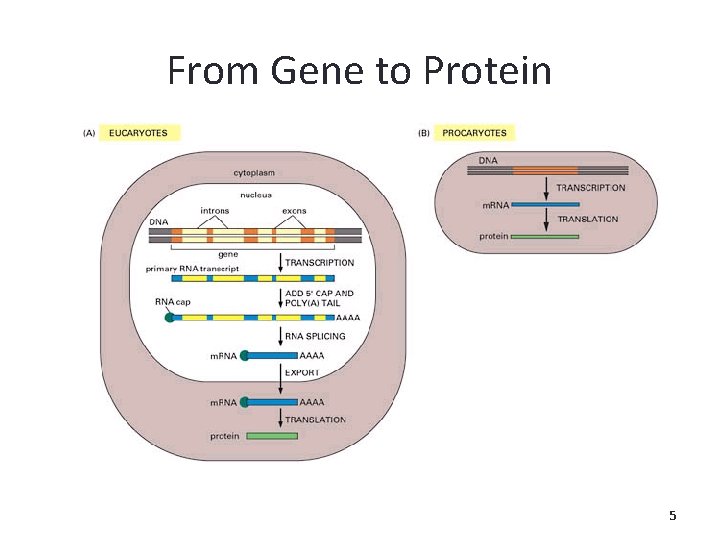
From Gene to Protein 5
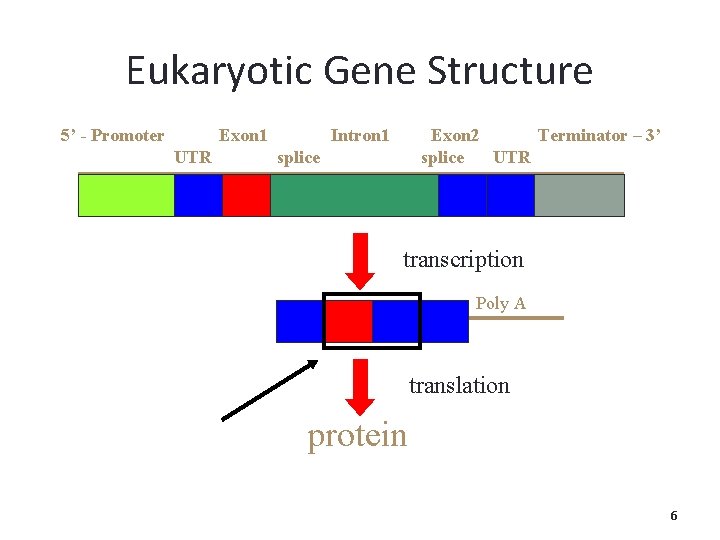
Eukaryotic Gene Structure 5’ - Promoter Exon 1 UTR Intron 1 Exon 2 Terminator – 3’ splice UTR splice transcription Poly A translation protein 6
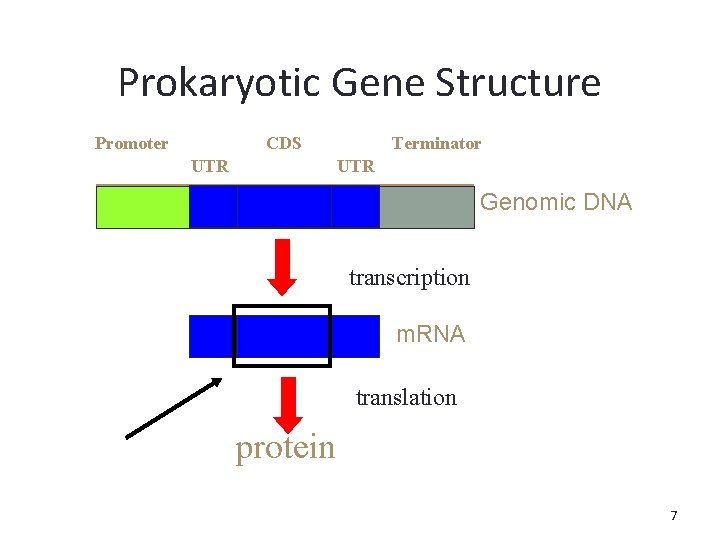
Prokaryotic Gene Structure Promoter CDS UTR Terminator UTR Genomic DNA transcription m. RNA translation protein 7
Topics to be Covered • Introduction • Gene structure – Promoter – Terminator – Splice sites 8

Promoter • Promoter determines: 1. Which strand will serve as a template. 2. Transcription starting point. 3. Strength of polymerase binding. 4. Frequency of polymerase binding. 9
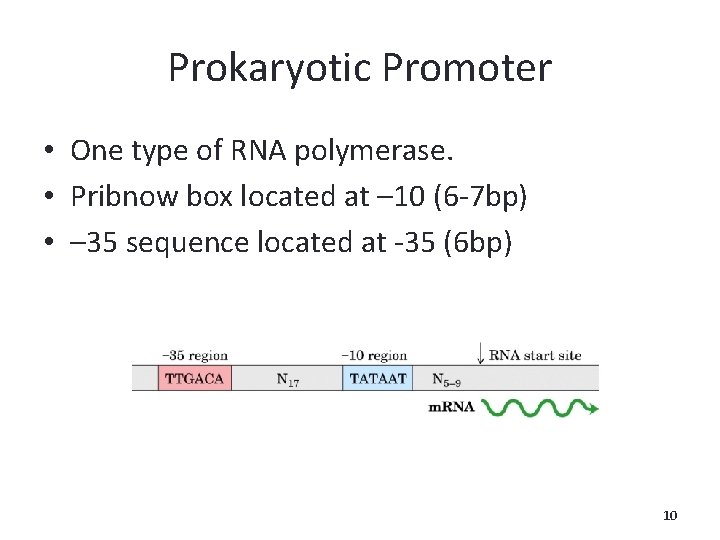
Prokaryotic Promoter • One type of RNA polymerase. • Pribnow box located at – 10 (6 -7 bp) • – 35 sequence located at -35 (6 bp) 10
Eukaryote Promoter • 3 types of RNA polymerases are employed in transcription of genes: – RNA polymerase I transcribes r. RNA – RNA polymerase II transcribes all genes coding for polypeptides – RNA polymerase III transcribes small cytoplasmatic RNA, such as t. RNA. 11
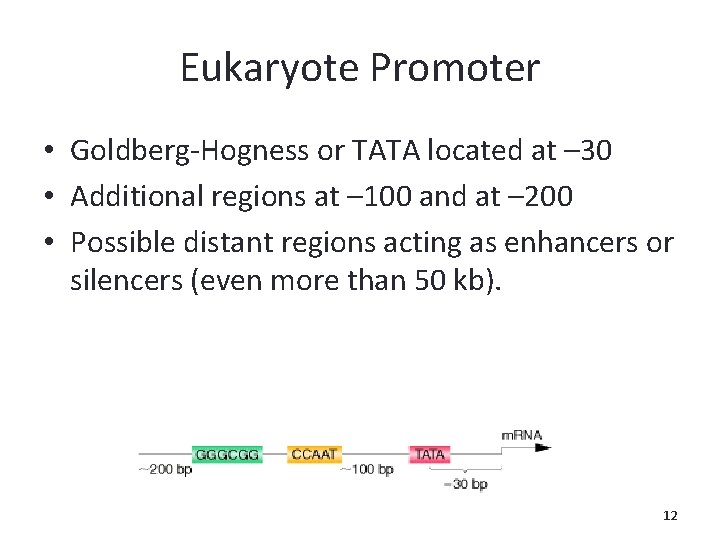
Eukaryote Promoter • Goldberg-Hogness or TATA located at – 30 • Additional regions at – 100 and at – 200 • Possible distant regions acting as enhancers or silencers (even more than 50 kb). 12
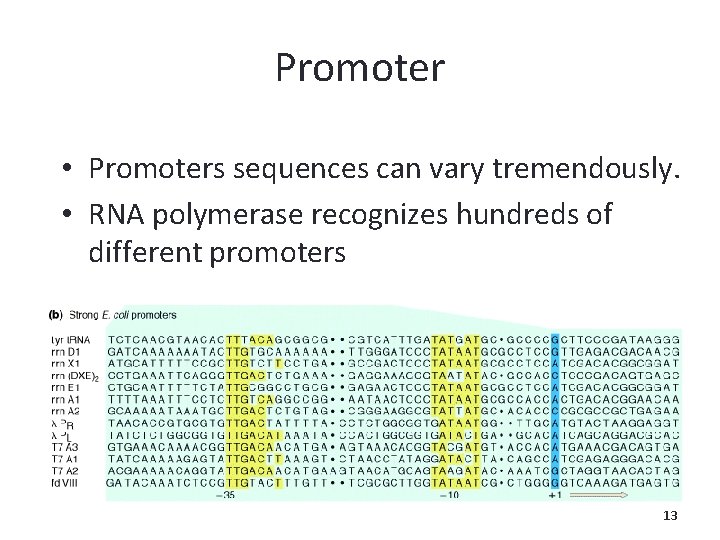
Promoter • Promoters sequences can vary tremendously. • RNA polymerase recognizes hundreds of different promoters 13
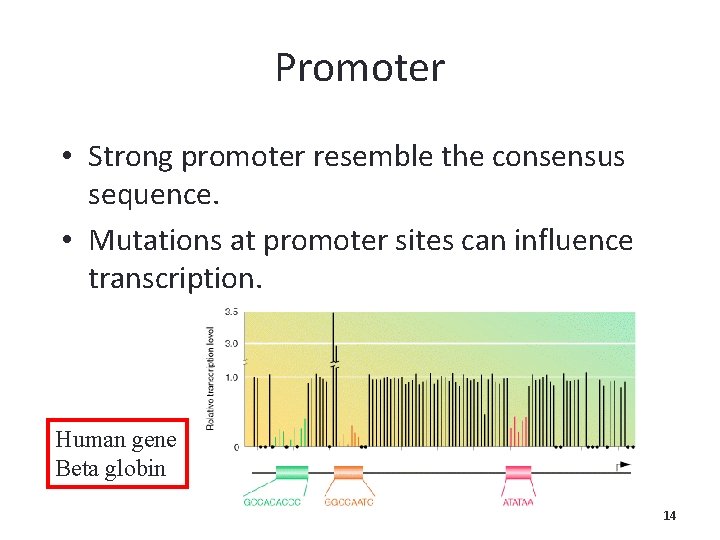
Promoter • Strong promoter resemble the consensus sequence. • Mutations at promoter sites can influence transcription. Human gene Beta globin 14

Promoter • Conclusions: 1. Promoters are very hard to predict. 2. Promoter prediction must be organismdependent (and even polymerase-dependent). 15
Termination Sites • The newly synthesized m. RNA forms a stem and loop structure (lollipop). • A disassociation signal at the end of the gene that stops elongating and releases RNA polymerase. • All terminators (eukaryotes and prokaryotes) form a secondary structure. 16
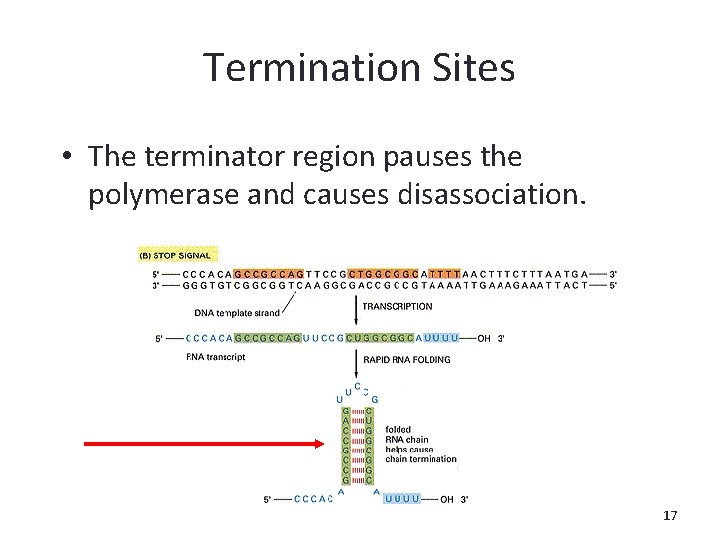
Termination Sites • The terminator region pauses the polymerase and causes disassociation. 17
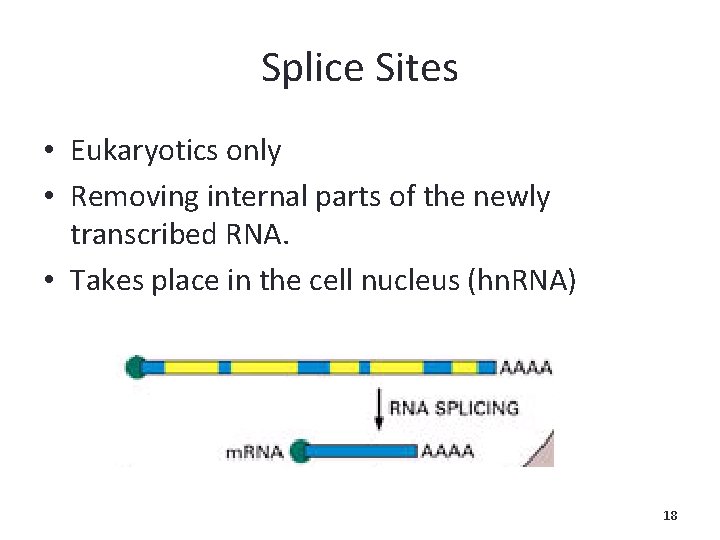
Splice Sites • Eukaryotics only • Removing internal parts of the newly transcribed RNA. • Takes place in the cell nucleus (hn. RNA) 18
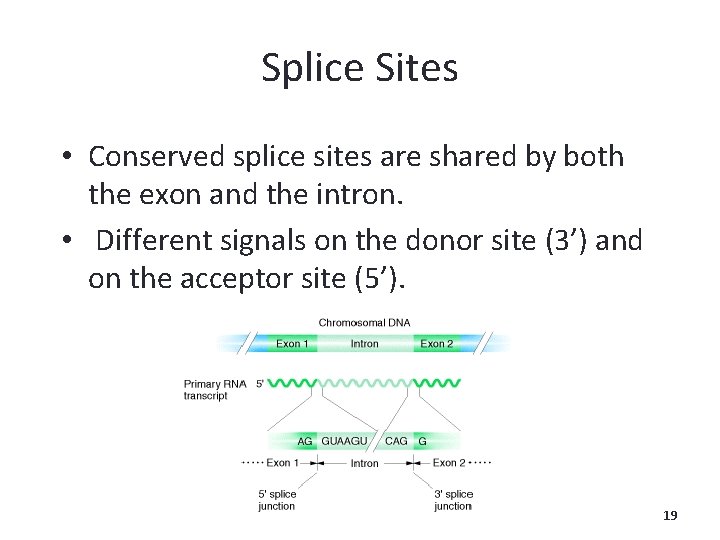
Splice Sites • Conserved splice sites are shared by both the exon and the intron. • Different signals on the donor site (3’) and on the acceptor site (5’). 19
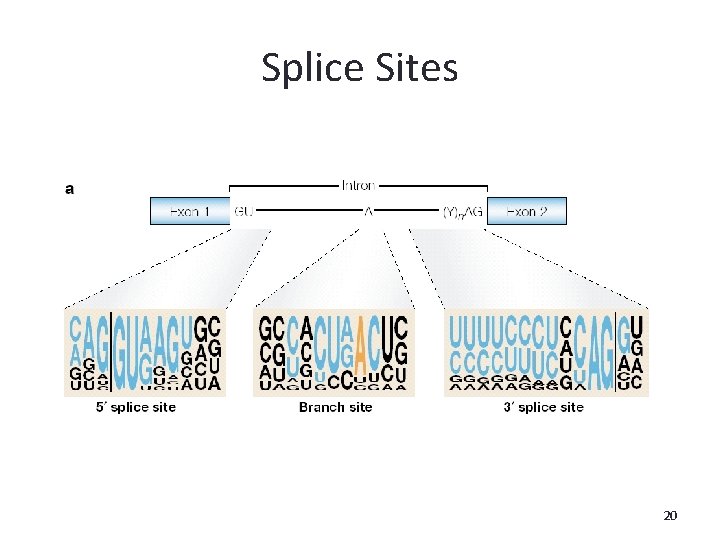
Splice Sites 20
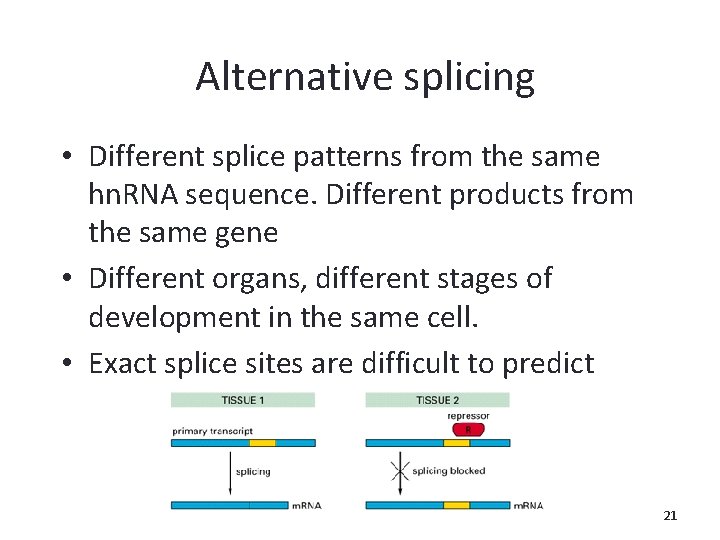
Alternative splicing • Different splice patterns from the same hn. RNA sequence. Different products from the same gene • Different organs, different stages of development in the same cell. • Exact splice sites are difficult to predict 21
- Slides: 21