Gene Regulation Activation Repression Why Regulate 1 Control






- Slides: 65

Gene Regulation: Activation & Repression

Why Regulate? 1. Control the quantities of gene products • Some are needed in large quantities: Ribosomes • Some are needed in small quantities: Regulatory proteins, Some enzymes. 2. Synthesize when the need arises • Respond to environment • Heat shock, UV radiation - Stress • New carbon and energy source • Quorum Sensing • When there is enough cell to make a difference 3. Need for temporal expression • Phage development Internal flexibility Provides adaptability

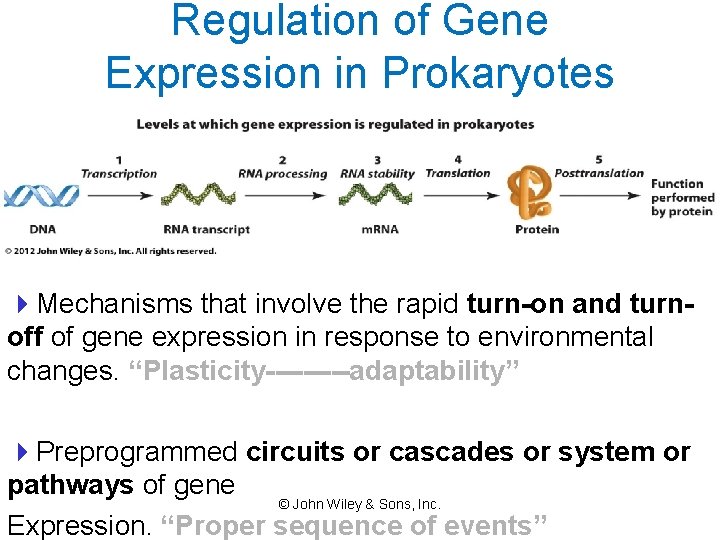
Regulation of Gene Expression in Prokaryotes 4 Mechanisms that involve the rapid turn-on and turnoff of gene expression in response to environmental changes. “Plasticity-----adaptability” 4 Preprogrammed circuits or cascades or system or pathways of gene © John Wiley & Sons, Inc. Expression. “Proper sequence of events”


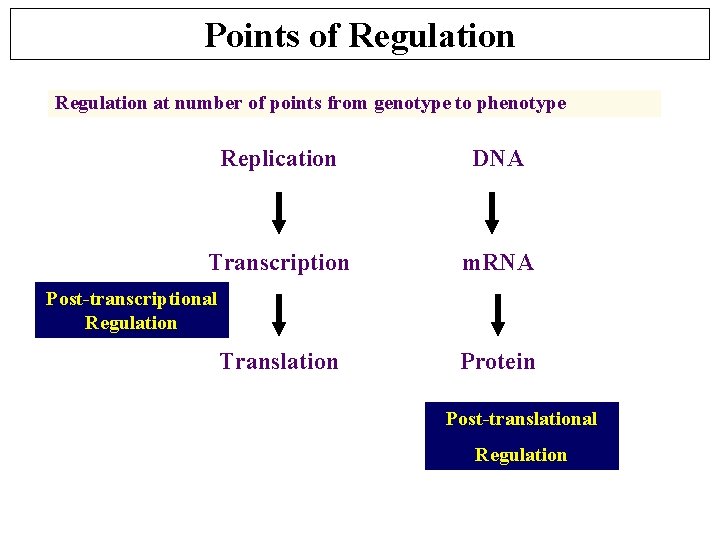
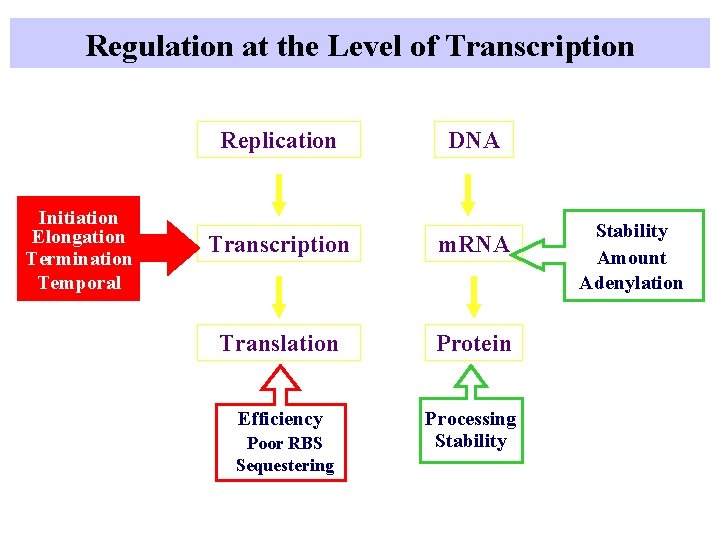
Points of Regulation at number of points from genotype to phenotype Replication DNA Transcription m. RNA Post-transcriptional Regulation Translation Protein Post-translational Regulation

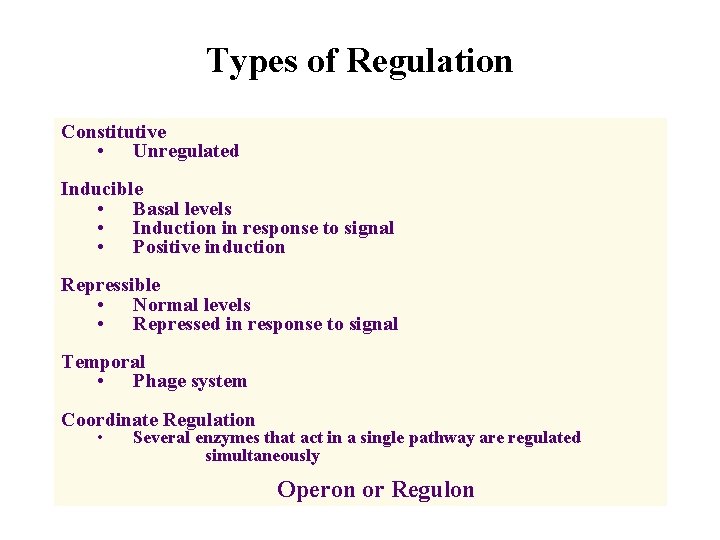
Types of Regulation Constitutive • Unregulated Inducible • Basal levels • Induction in response to signal • Positive induction Repressible • Normal levels • Repressed in response to signal Temporal • Phage system Coordinate Regulation • Several enzymes that act in a single pathway are regulated simultaneously Operon or Regulon

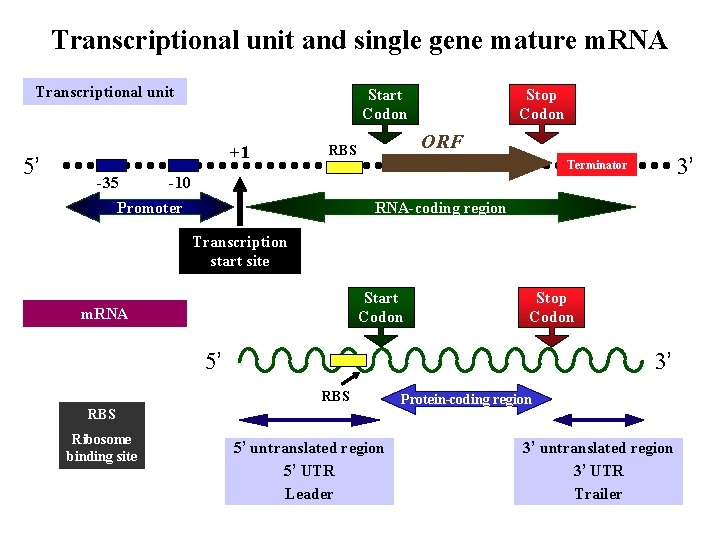
Transcriptional unit and single gene mature m. RNA Transcriptional unit 5’ Start Codon +1 -35 Stop Codon ORF RBS 3’ Terminator -10 Promoter RNA-coding region Transcription start site Start Codon m. RNA Stop Codon 5’ 3’ RBS Ribosome binding site 5’ untranslated region 5’ UTR Leader Protein-coding region 3’ untranslated region 3’ UTR Trailer

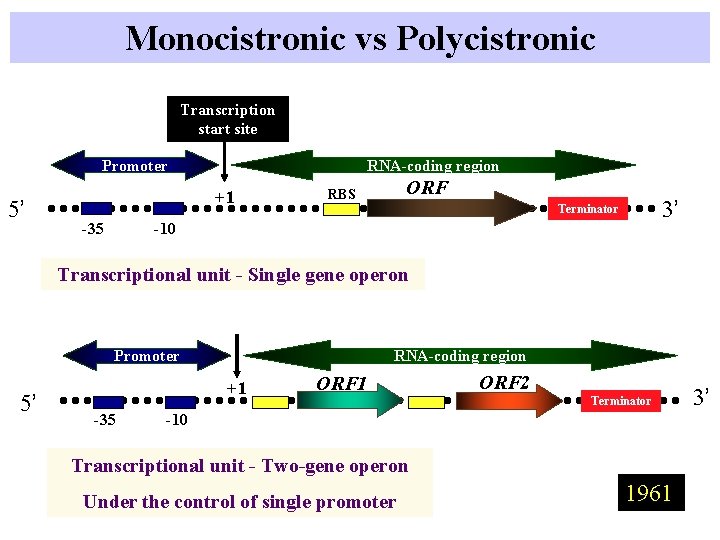
Monocistronic vs Polycistronic Transcription start site Promoter 5’ RNA-coding region +1 -35 ORF RBS 3’ Terminator -10 Transcriptional unit - Single gene operon Promoter 5’ RNA-coding region +1 -35 ORF 1 ORF 2 Terminator -10 Transcriptional unit - Two-gene operon Under the control of single promoter 1961 3’

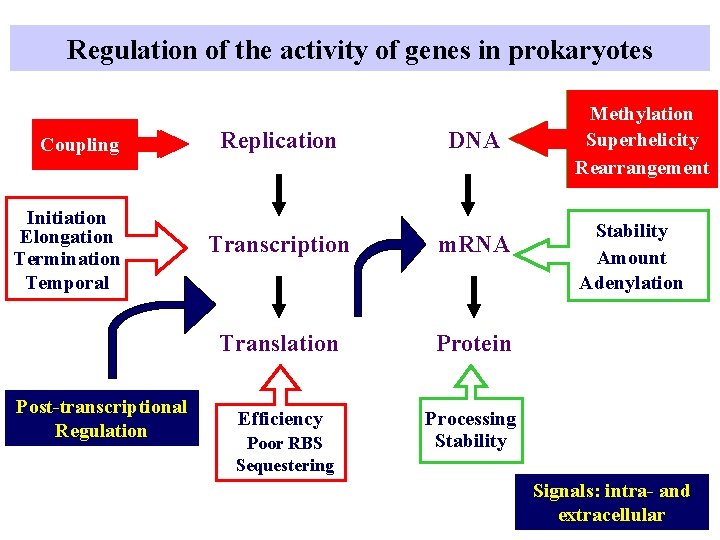
Regulation of the activity of genes in prokaryotes Coupling Initiation Elongation Termination Temporal Post-transcriptional Regulation Replication DNA Transcription m. RNA Translation Protein Efficiency Processing Stability Poor RBS Sequestering Methylation Superhelicity Rearrangement Stability Amount Adenylation Signals: intra- and extracellular

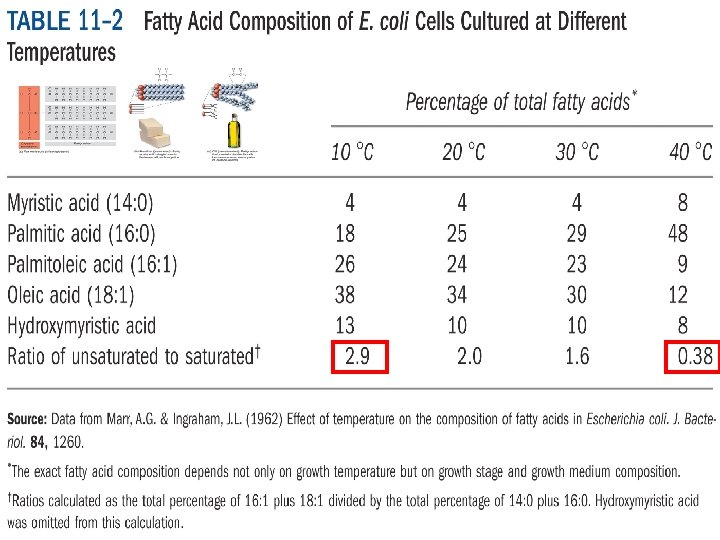
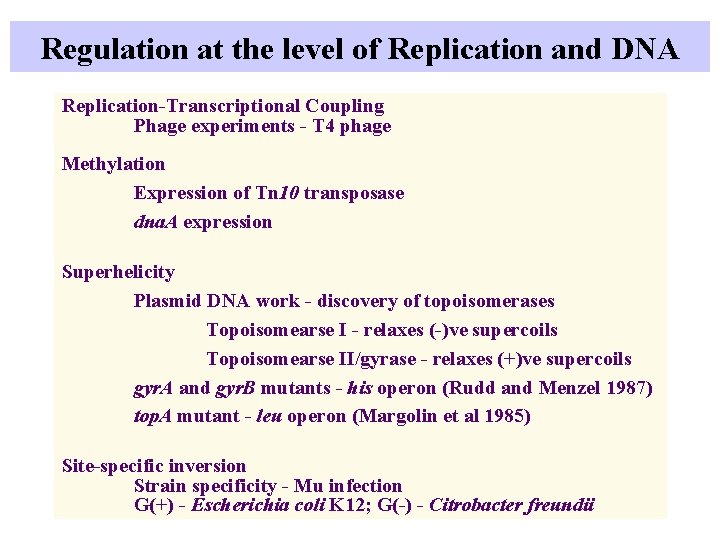
Regulation at the level of Replication and DNA Replication-Transcriptional Coupling Phage experiments - T 4 phage Methylation Expression of Tn 10 transposase dna. A expression Superhelicity Plasmid DNA work - discovery of topoisomerases Topoisomearse I - relaxes (-)ve supercoils Topoisomearse II/gyrase - relaxes (+)ve supercoils gyr. A and gyr. B mutants - his operon (Rudd and Menzel 1987) top. A mutant - leu operon (Margolin et al 1985) Site-specific inversion Strain specificity - Mu infection G(+) - Escherichia coli K 12; G(-) - Citrobacter freundii

Regulation at the Level of Transcription Initiation Elongation Termination Temporal Replication DNA Transcription m. RNA Translation Protein Efficiency Processing Stability Poor RBS Sequestering Stability Amount Adenylation

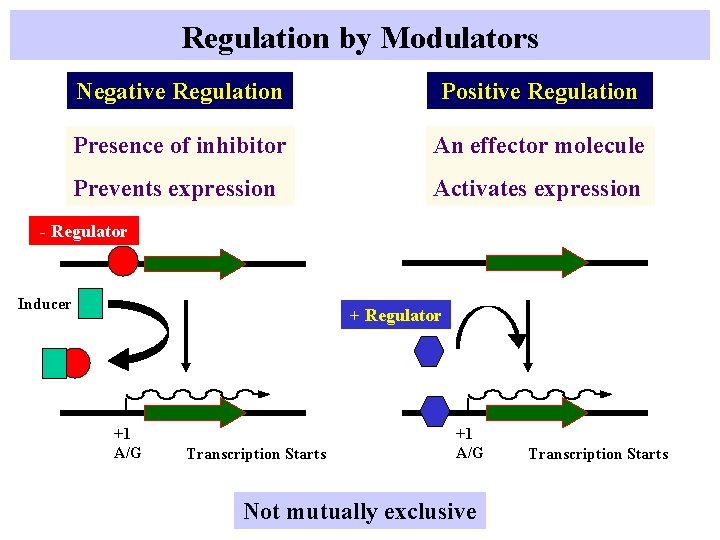
Regulation by Modulators Negative Regulation Positive Regulation Presence of inhibitor An effector molecule Prevents expression Activates expression - Regulator Inducer + Regulator +1 A/G Transcription Starts +1 A/G Not mutually exclusive Transcription Starts

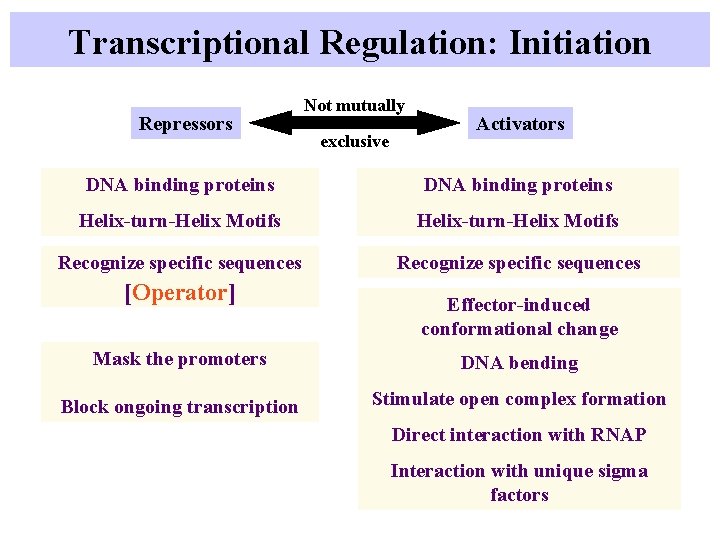
Transcriptional Regulation: Initiation Repressors Not mutually exclusive Activators DNA binding proteins Helix-turn-Helix Motifs Recognize specific sequences [Operator] Effector-induced conformational change Mask the promoters DNA bending Block ongoing transcription Stimulate open complex formation Direct interaction with RNAP Interaction with unique sigma factors

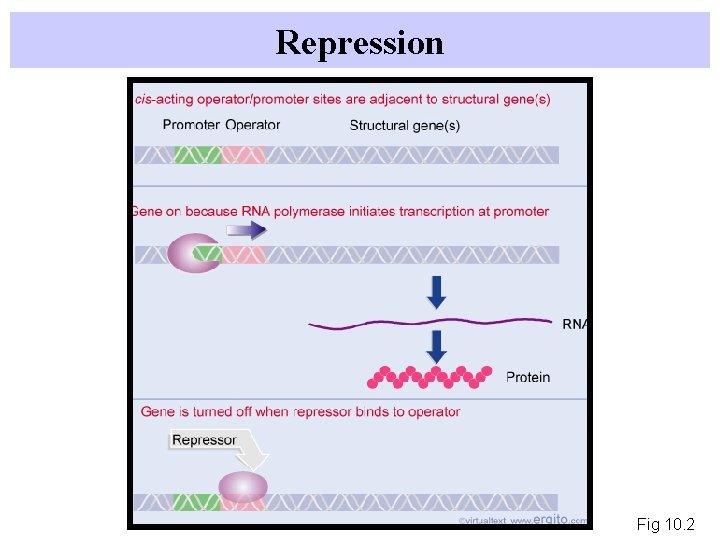
Repression Fig 10. 2

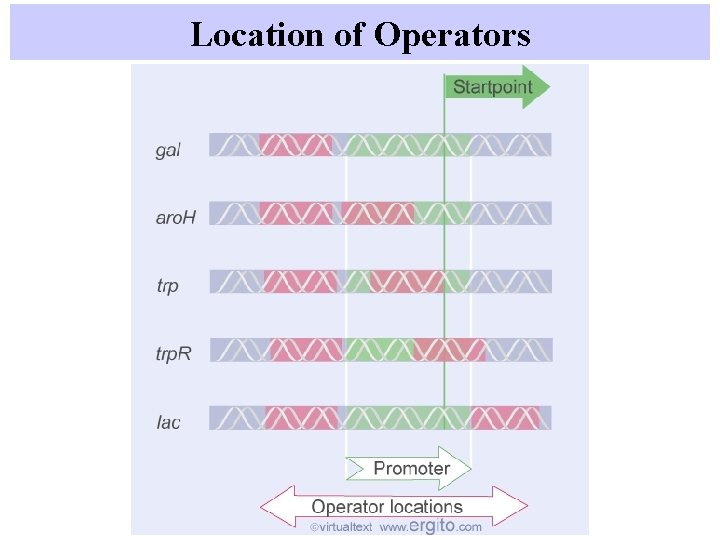
Location of Operators

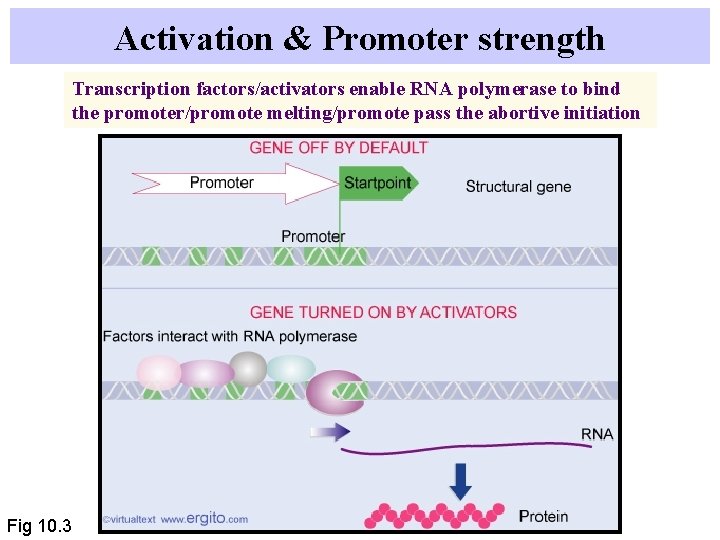
Activation & Promoter strength Transcription factors/activators enable RNA polymerase to bind the promoter/promote melting/promote pass the abortive initiation Fig 10. 3

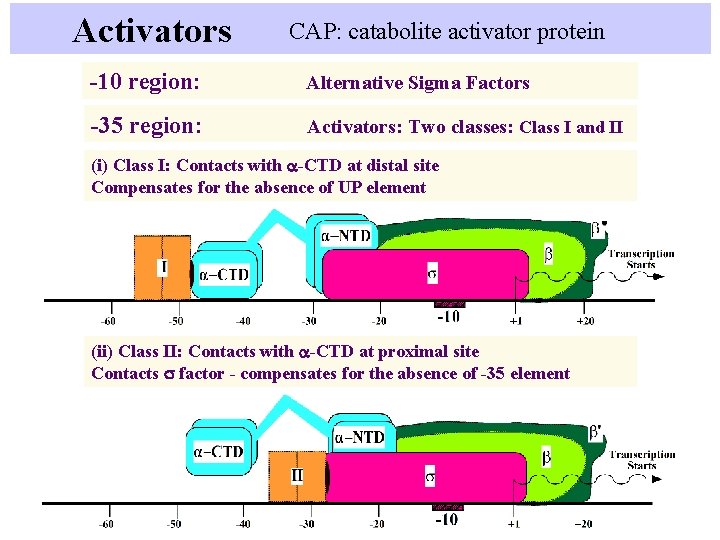
Activators CAP: catabolite activator protein -10 region: Alternative Sigma Factors -35 region: Activators: Two classes: Class I and II (i) Class I: Contacts with a-CTD at distal site Compensates for the absence of UP element (ii) Class II: Contacts with a-CTD at proximal site Contacts s factor - compensates for the absence of -35 element

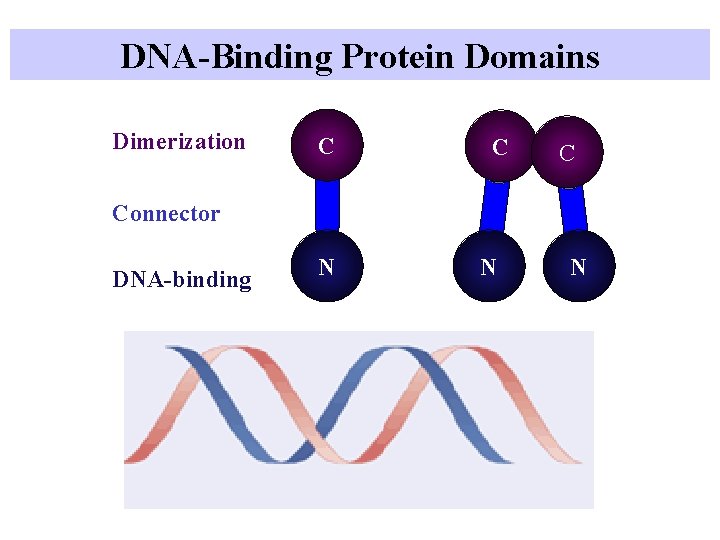
DNA-Binding Protein Domains Dimerization C Connector DNA-binding N N N

DNA-Binding Motifs Helix-turn-Helix (HTH) Two alpha helices connected by a turn Helix-loop-Helix (HLH) Two alpha helices connected by a loop Eukaryotic system Homeodomain: Three helices (almost resembles HTH) Zinc Fingers Steroid Receptors Leucine Zippers

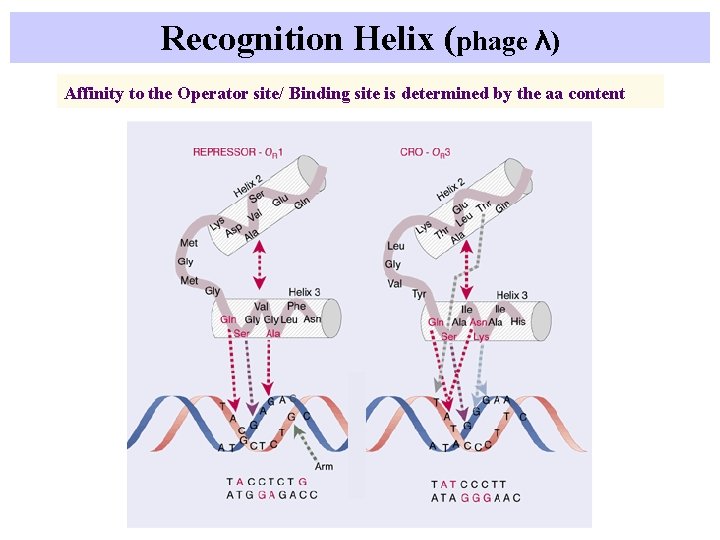
Recognition Helix (phage λ) Affinity to the Operator site/ Binding site is determined by the aa content

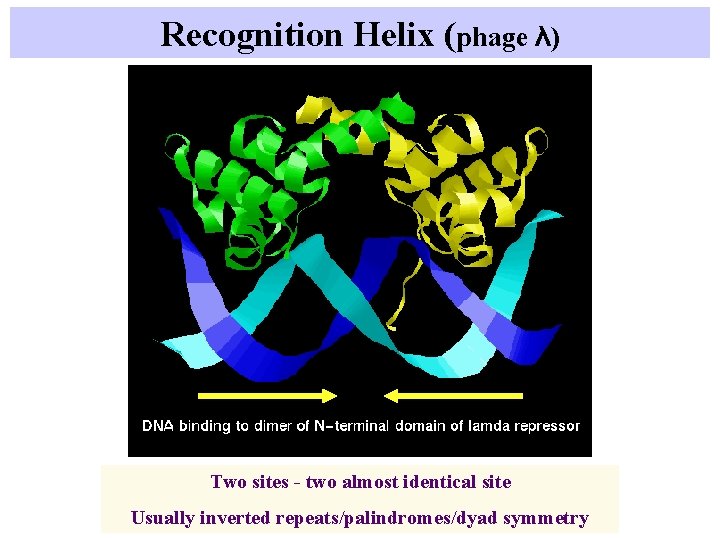
Recognition Helix (phage λ) Two sites - two almost identical site Usually inverted repeats/palindromes/dyad symmetry

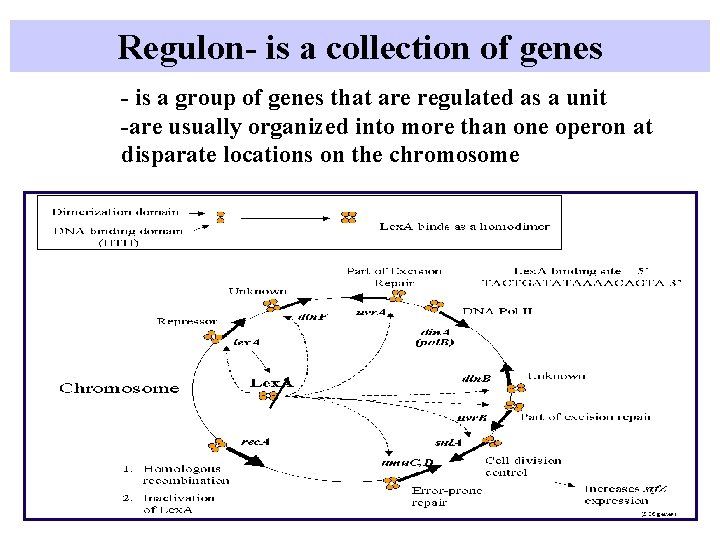
Regulon- is a collection of genes - is a group of genes that are regulated as a unit -are usually organized into more than one operon at disparate locations on the chromosome

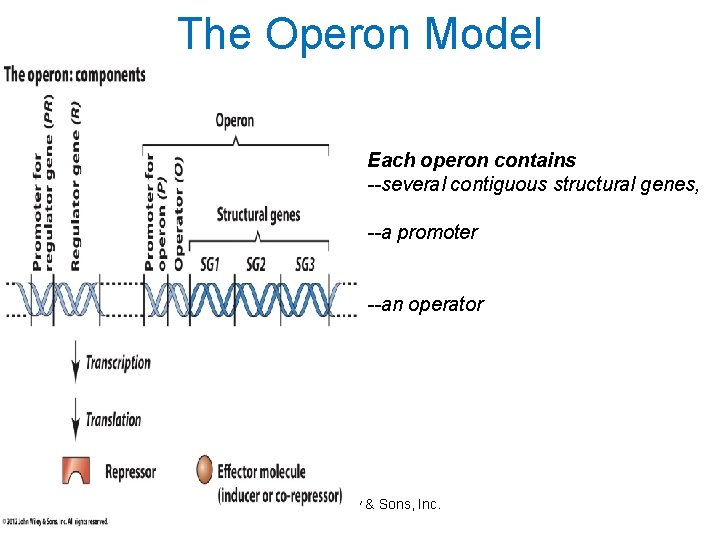
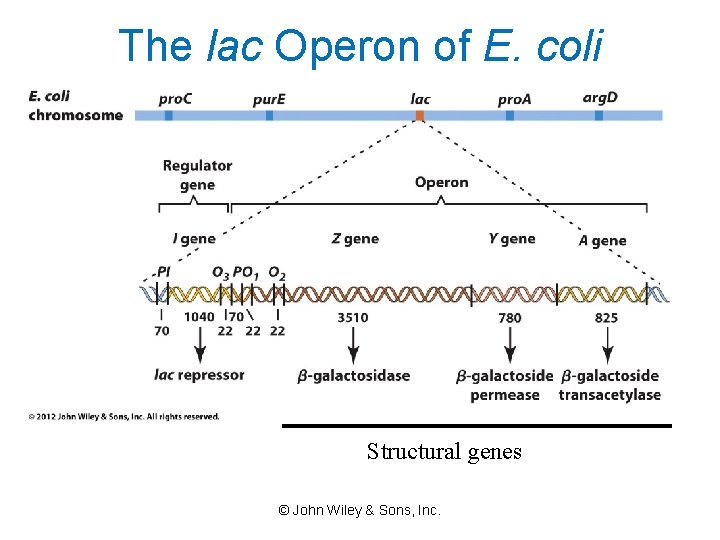
Operons: Units required for Gene Expression Sequence of DNA In prokaryotes, the operon includes structural genes, the operator and the promoter. © John Wiley & Sons, Inc.

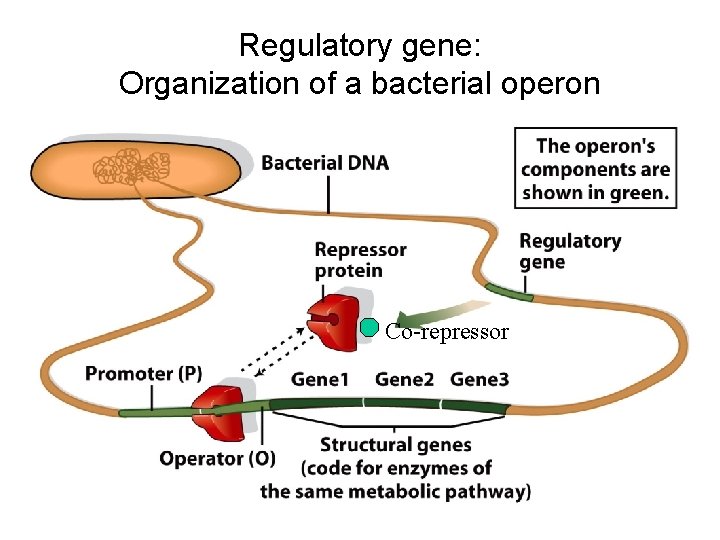
Regulatory gene: Organization of a bacterial operon

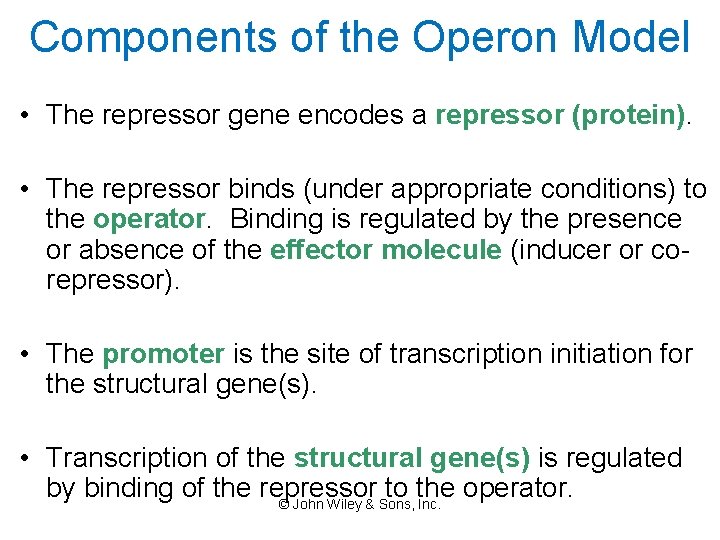
Components of the Operon Model • The repressor gene encodes a repressor (protein). • The repressor binds (under appropriate conditions) to the operator. Binding is regulated by the presence or absence of the effector molecule (inducer or corepressor). • The promoter is the site of transcription initiation for the structural gene(s). • Transcription of the structural gene(s) is regulated by binding of the repressor to the operator. © John Wiley & Sons, Inc.

Regulatory gene: Organization of a bacterial operon Co-repressor

Control of Gene Expression in Bacteria • The Bacterial Operon – An operon is a functional complex of genes containing the information for enzymes of a metabolic pathway. It includes: • Structural genes – code for the enzymes and are translated from a single m. RNA (Polycistronic). • Promoter – where the RNA polymerase binds. • Operator – site next to the promoter , where the regulatory protein can bind. • A repressor (~proteins) which binds to a specific DNA sequence to determine whether or not a particular gene is transcribed. • The regulatory gene encodes the repressor protein

The Operon Model Each operon contains --several contiguous structural genes, --a promoter --an operator © John Wiley & Sons, Inc.

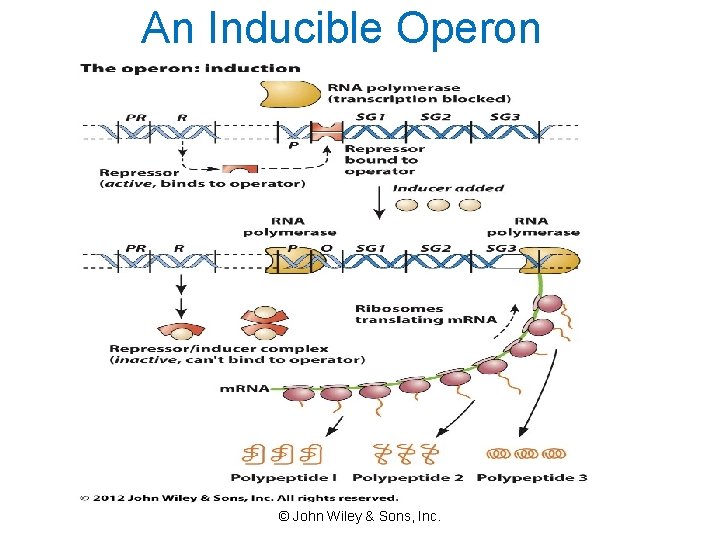
An Inducible Operon © John Wiley & Sons, Inc.

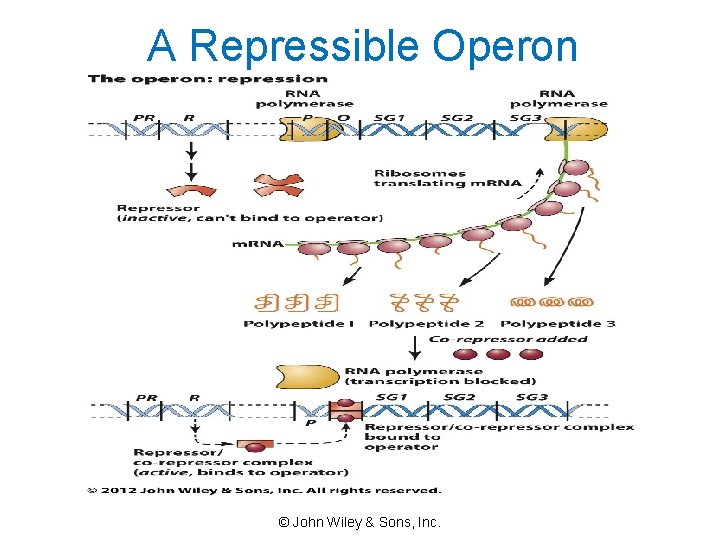
A Repressible Operon © John Wiley & Sons, Inc.

The Structural Genes of an Operon • A single m. RNA transcript carries the coding information of an entire operon. • Operons containing more than one structural gene are multigenic. • All structural genes in an operon are co-transcribed and therefore are coordinately expressed. © John Wiley & Sons, Inc.

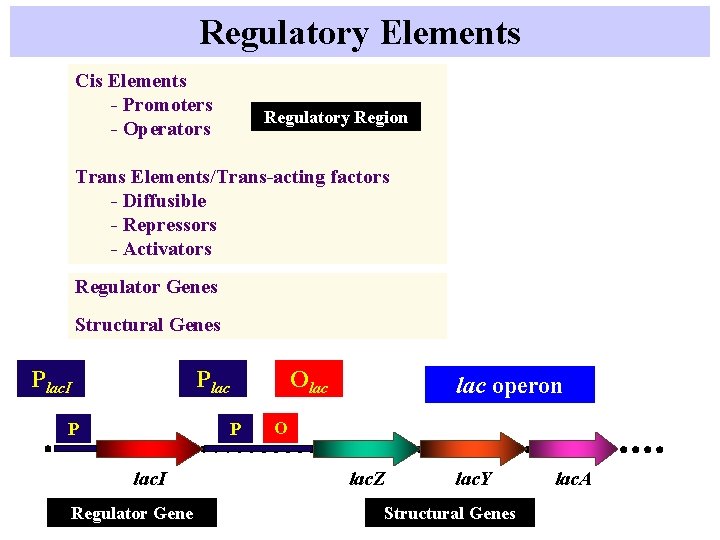
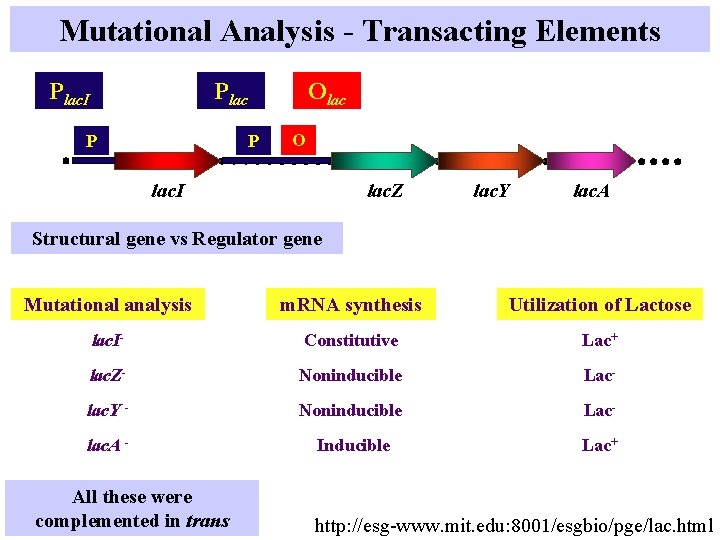
Regulatory Elements Cis Elements - Promoters - Operators Regulatory Region Trans Elements/Trans-acting factors - Diffusible - Repressors - Activators Regulator Genes Structural Genes Plac. I Olac P P lac. I Regulator Gene lac operon O lac. Z lac. Y Structural Genes lac. A

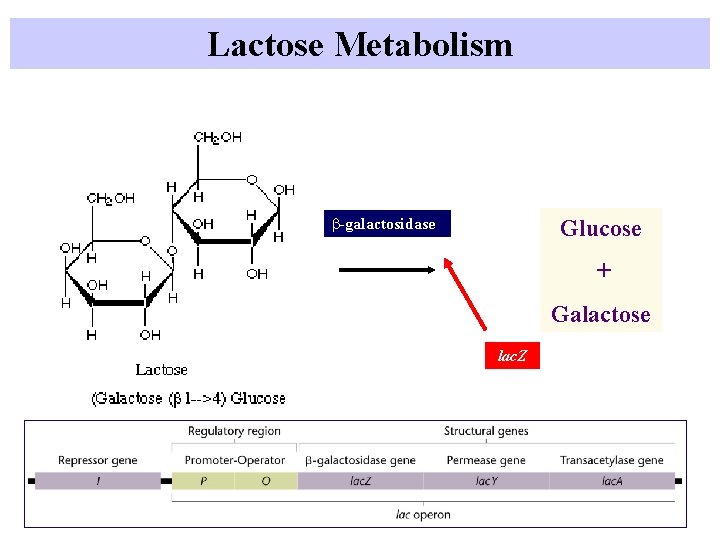
Lactose Metabolism b-galactosidase Glucose + Galactose lac. Z

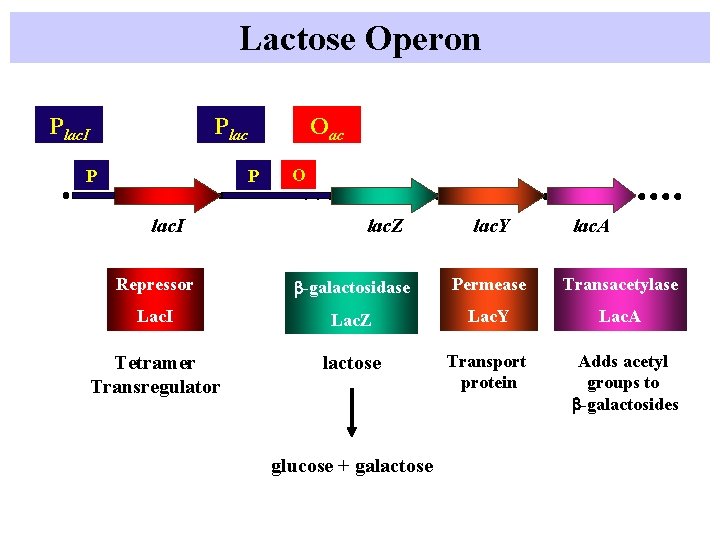
Lactose Operon Plac. I Oac P P lac. I O lac. Z lac. Y lac. A Repressor b-galactosidase Permease Transacetylase Lac. I Lac. Z Lac. Y Lac. A Tetramer Transregulator lactose Transport protein Adds acetyl groups to b-galactosides glucose + galactose

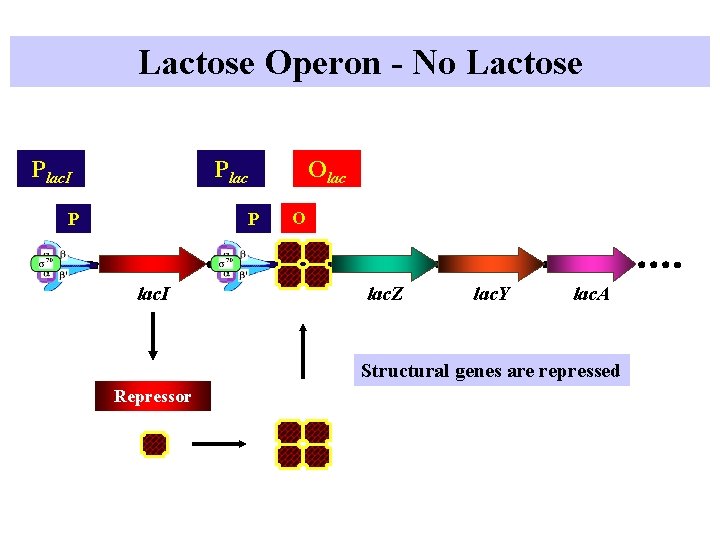
Lactose Operon - No Lactose Plac. I Plac P Olac P lac. I O lac. Z lac. Y lac. A Structural genes are repressed Repressor

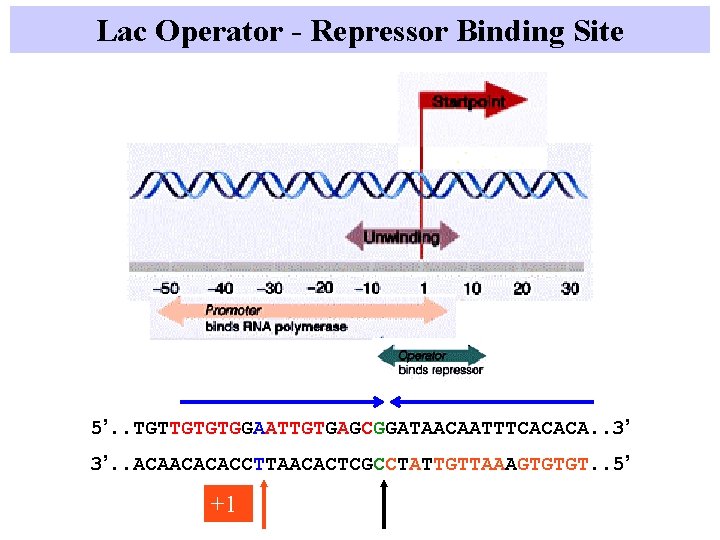
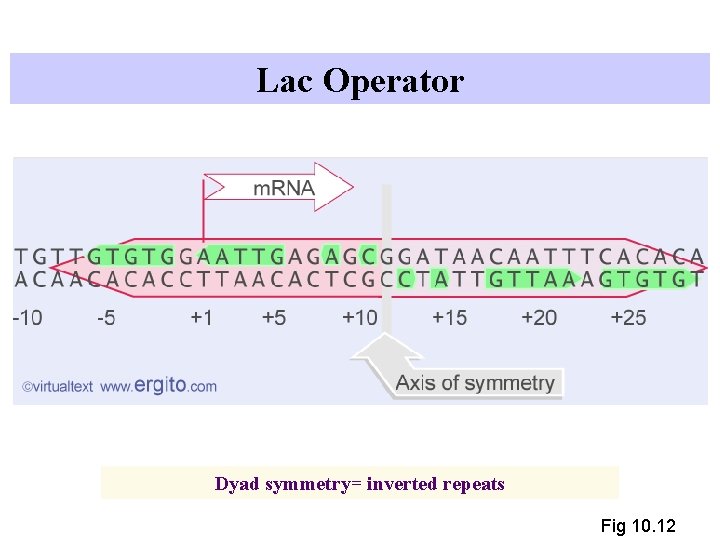
Lac Operator - Repressor Binding Site 5’. . TGTTGTGTGGAATTGTGAGCGGATAACAATTTCACACA. . 3’ 3’. . ACAACACACCTTAACACTCGCCTATTGTTAAAGTGTGT. . 5’ +1

Lac Operator Dyad symmetry= inverted repeats Fig 10. 12

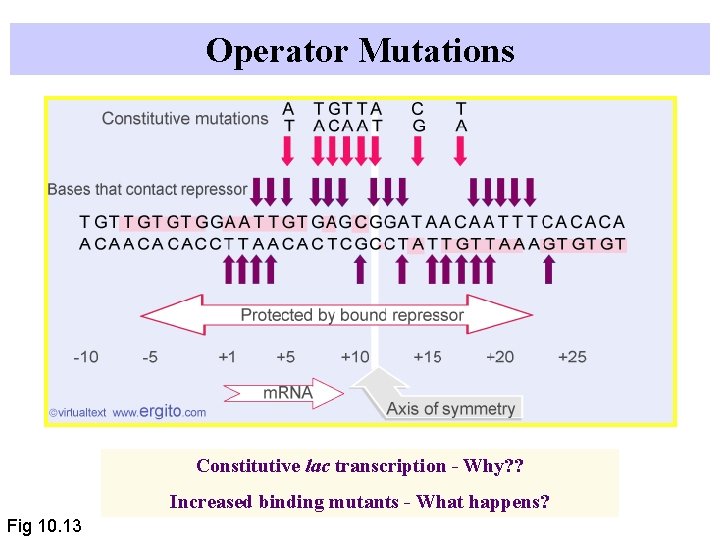
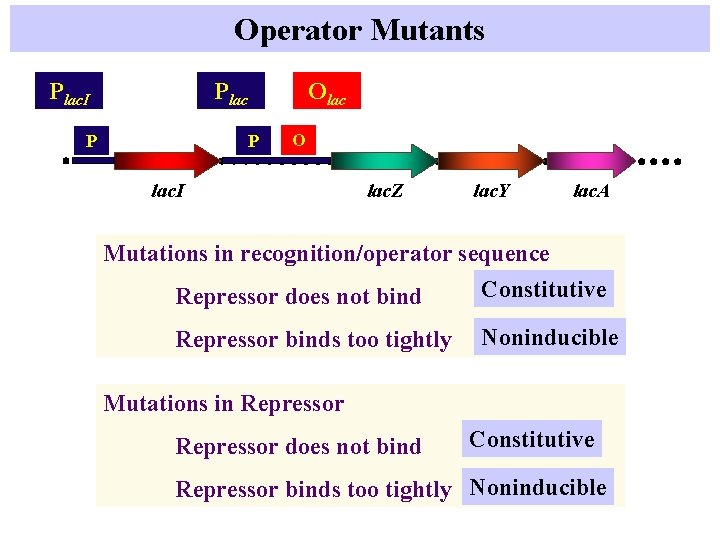
Operator Mutations Constitutive lac transcription - Why? ? Increased binding mutants - What happens? Fig 10. 13

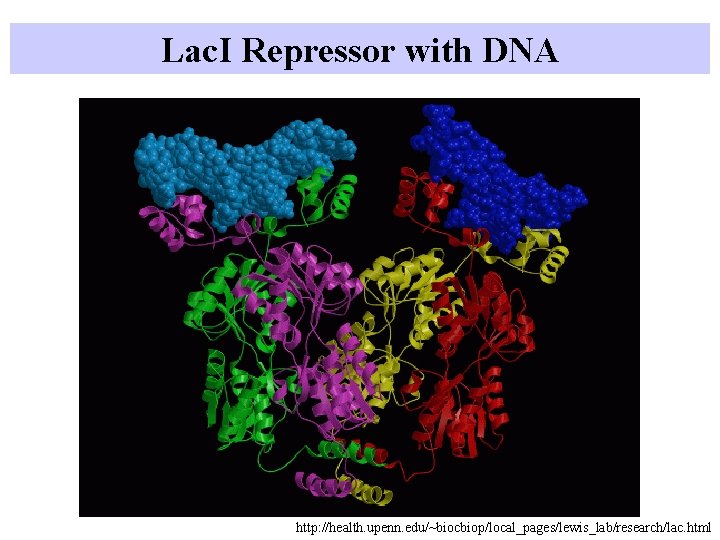
Lac. I Repressor with DNA http: //health. upenn. edu/~biocbiop/local_pages/lewis_lab/research/lac. html

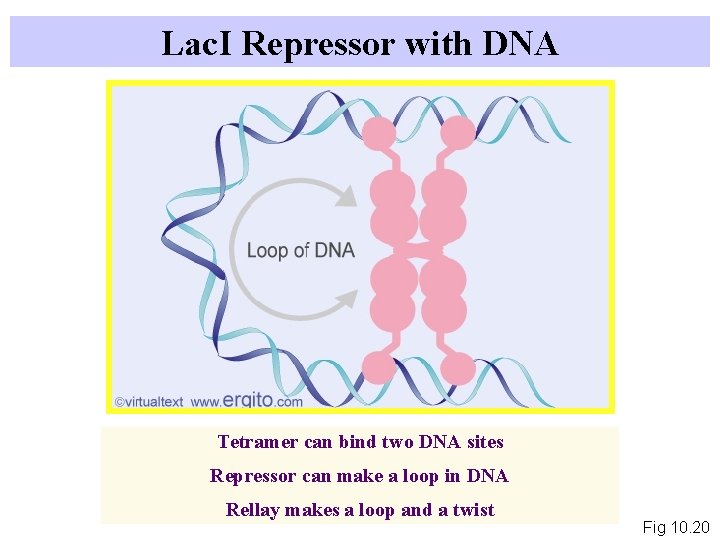
Lac. I Repressor with DNA Tetramer can bind two DNA sites Repressor can make a loop in DNA Rellay makes a loop and a twist Fig 10. 20

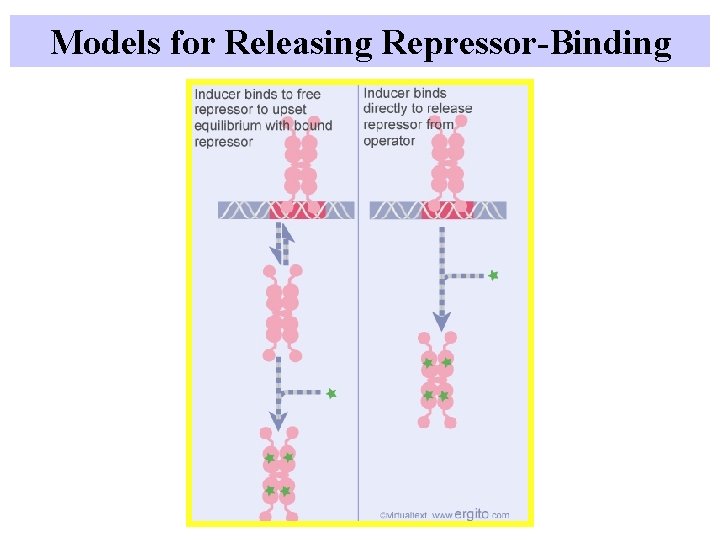
Models for Releasing Repressor-Binding

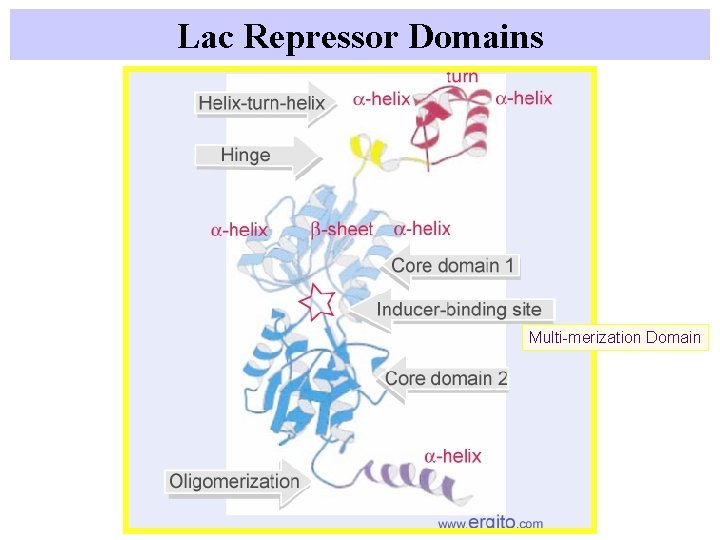
Lac Repressor Domains Multi-merization Domain

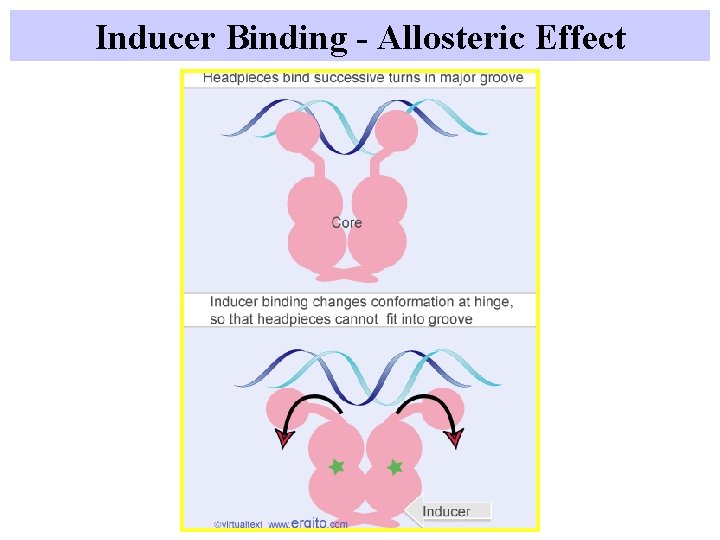
Inducer Binding - Allosteric Effect

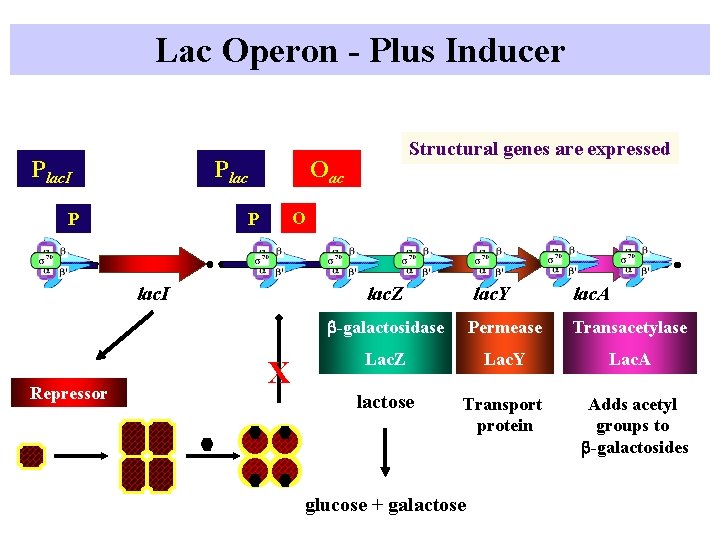
Lac Operon - Plus Inducer Plac. I Plac P Oac O P lac. I Repressor Structural genes are expressed lac. Z X lac. Y lac. A b-galactosidase Permease Transacetylase Lac. Z Lac. Y Lac. A lactose Transport protein Adds acetyl groups to b-galactosides glucose + galactose

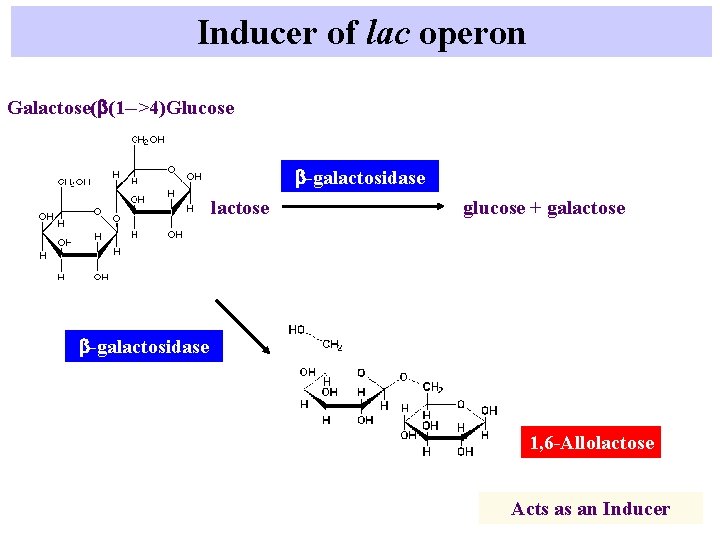
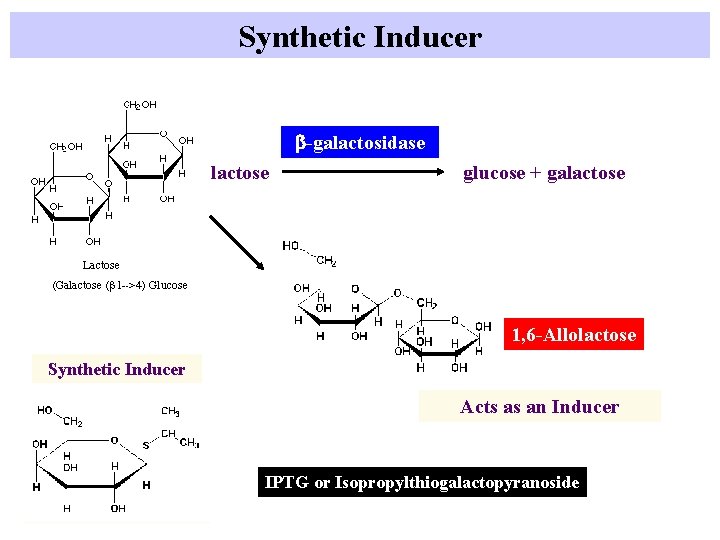
Inducer of lac operon Galactose(b(1 -->4)Glucose b-galactosidase lactose glucose + galactose b-galactosidase 1, 6 -Allolactose Acts as an Inducer

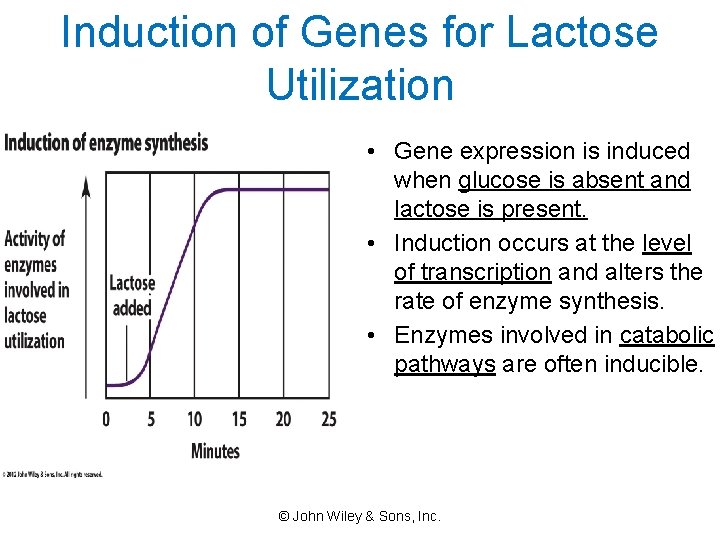
Induction of Genes for Lactose Utilization • Gene expression is induced when glucose is absent and lactose is present. • Induction occurs at the level of transcription and alters the rate of enzyme synthesis. • Enzymes involved in catabolic pathways are often inducible. © John Wiley & Sons, Inc.

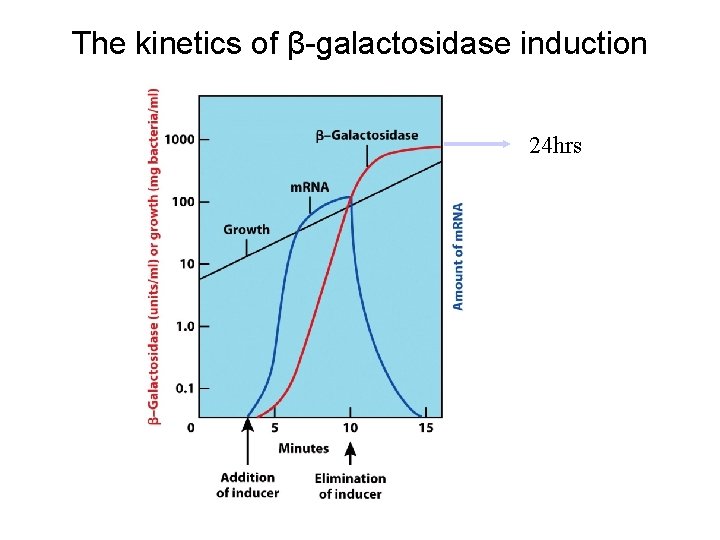
The kinetics of β-galactosidase induction 24 hrs

The lac Operon of E. coli Structural genes © John Wiley & Sons, Inc.

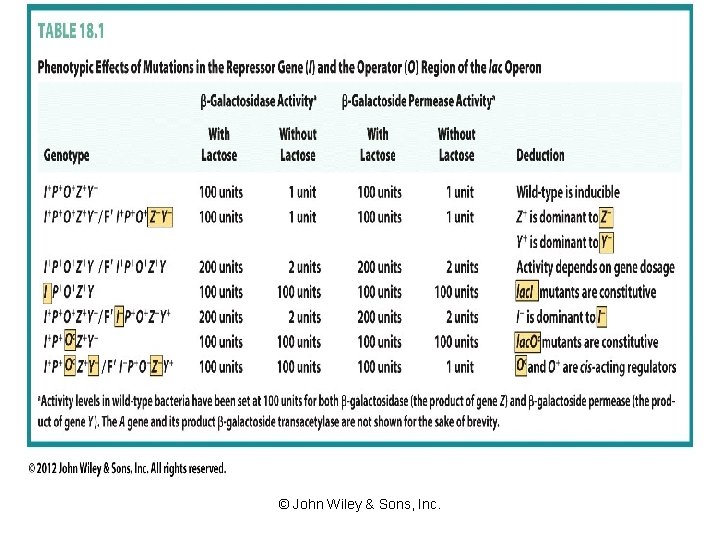
Mutational Analysis - Transacting Elements Plac. I Olac P P O lac. I lac. Z lac. Y lac. A Structural gene vs Regulator gene Mutational analysis m. RNA synthesis Utilization of Lactose lac. I- Constitutive Lac+ lac. Z- Noninducible Lac- lac. Y - Noninducible Lac- lac. A - Inducible Lac+ All these were complemented in trans http: //esg-www. mit. edu: 8001/esgbio/pge/lac. html

Operator Mutants Plac. I Olac P P O lac. I lac. Z lac. Y lac. A Mutations in recognition/operator sequence Constitutive Repressor does not bind Repressor binds too tightly Noninducible Mutations in Repressor does not bind Constitutive Repressor binds too tightly Noninducible

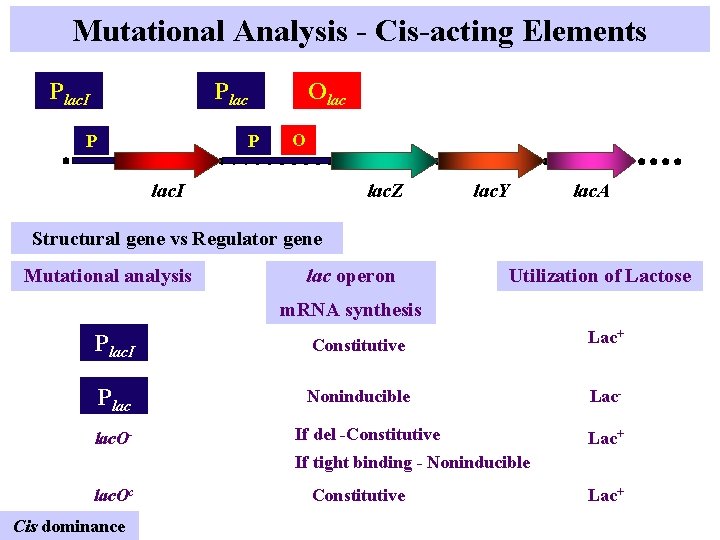
Mutational Analysis - Cis-acting Elements Plac. I Olac P P O lac. I lac. Z lac. Y lac. A Structural gene vs Regulator gene Mutational analysis lac operon Utilization of Lactose m. RNA synthesis Plac. I Constitutive Lac+ Plac Noninducible Lac- lac. O- If del -Constitutive Lac+ If tight binding - Noninducible lac. Oc Cis dominance Constitutive Lac+

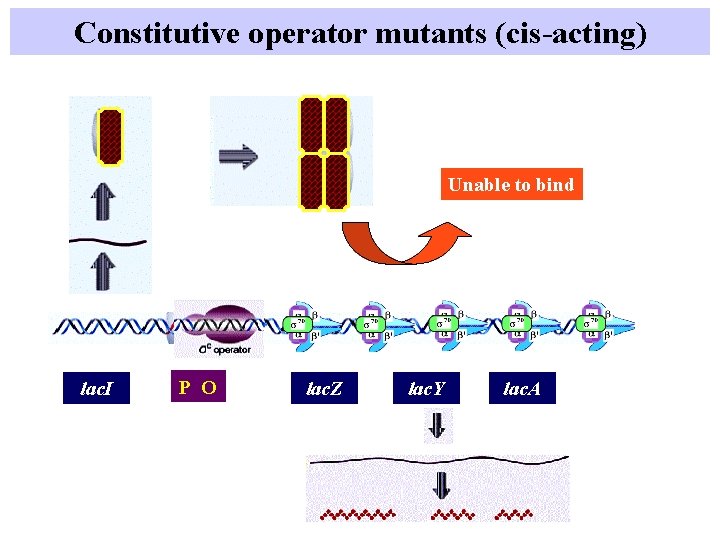
Constitutive operator mutants (cis-acting) Unable to bind lac. I P O lac. Z lac. Y lac. A

Mutations in the Structural Genes • The wild-type alleles (I+P+O+Z+Y+A+) of the structural genes are dominant to the mutant alleles (I-P-O-Z-Y-A-) • Genotypes F’ I+P+O+Z+Y+A+ / I+P+O+Z-Y-A- and F’ I+P+O+Z-Y-A-/ I+P+O+Z+Y+A+ are inducible for utilization of lactose. © John Wiley & Sons, Inc.

© John Wiley & Sons, Inc.

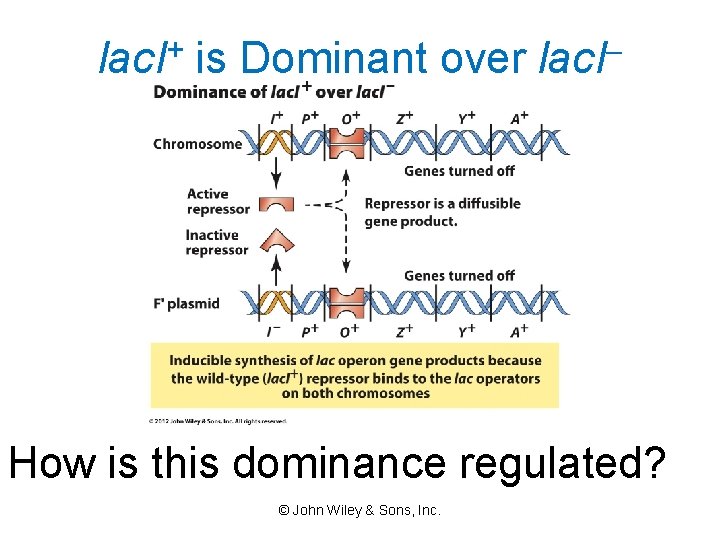
lac. I+ is Dominant over lac. I– How is this dominance regulated? © John Wiley & Sons, Inc.

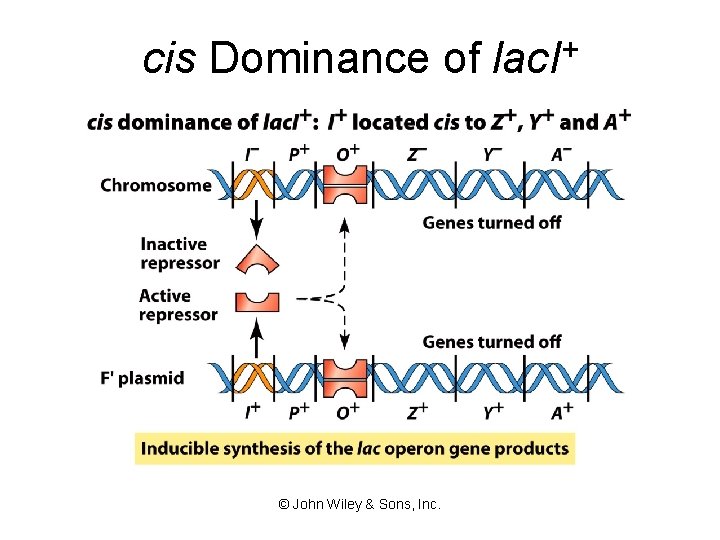
cis Dominance of lac. I+ © John Wiley & Sons, Inc.

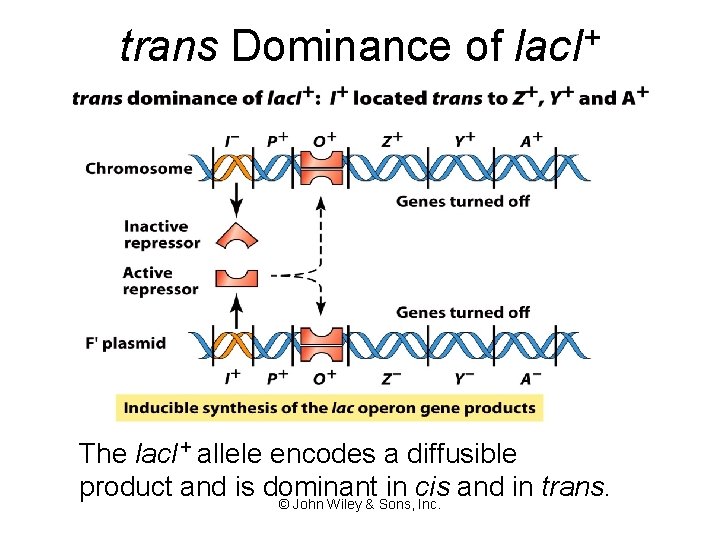
trans Dominance of lac. I+ The lac. I+ allele encodes a diffusible product and is dominant in cis and in trans. © John Wiley & Sons, Inc.

The operator • The Oc mutant is the binding site for the repressor and does not encode a product; this mutant acts only in cis. • Genotype F’ I+P+Oc. Z-Y-A- / I+P+O+Z+Y+A+ is inducible for the structural genes, but genotype F’ I+P+Oc. Z+Y+A+ / I+P+O+Z-Y-A- synthesizes the enzymes constitutively. © John Wiley & Sons, Inc.

© John Wiley & Sons, Inc.

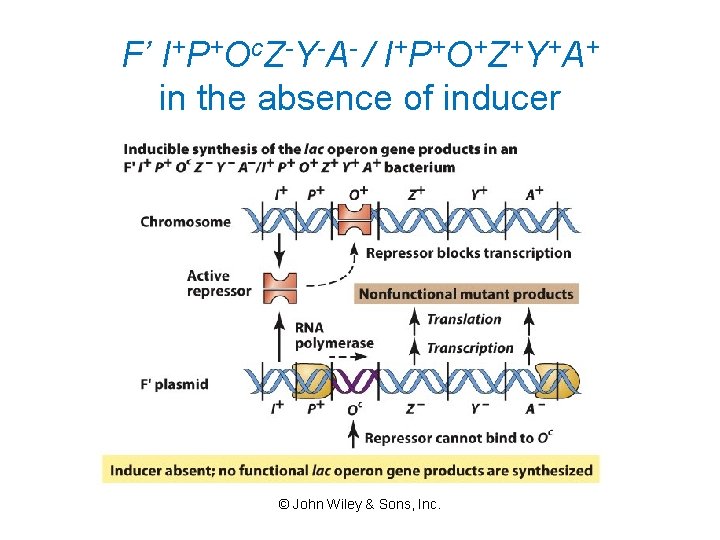
F’ I+P+Oc. Z-Y-A- / I+P+O+Z+Y+A+ in the absence of inducer © John Wiley & Sons, Inc.

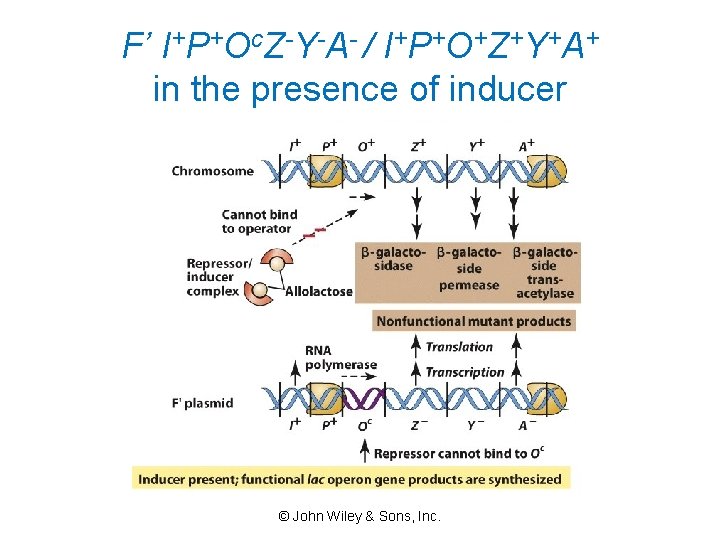
F’ I+P+Oc. Z-Y-A- / I+P+O+Z+Y+A+ in the presence of inducer © John Wiley & Sons, Inc.

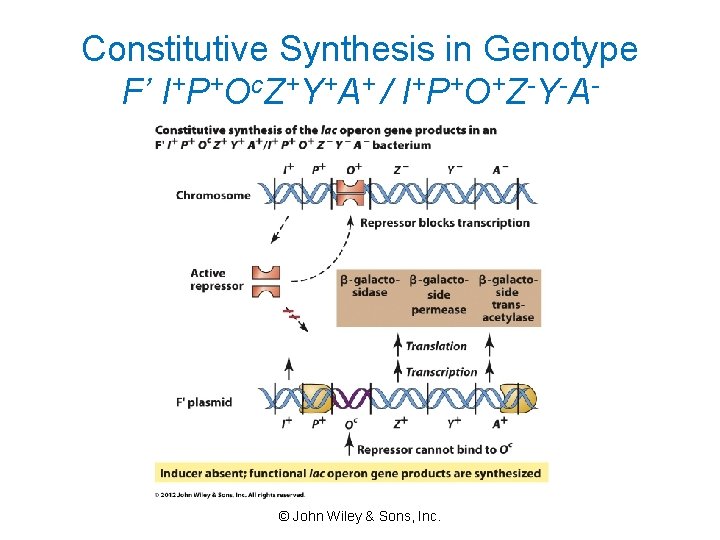
Constitutive Synthesis in Genotype F’ I+P+Oc. Z+Y+A+ / I+P+O+Z-Y-A- © John Wiley & Sons, Inc.

Synthetic Inducer b-galactosidase lactose glucose + galactose 1, 6 -Allolactose Synthetic Inducer Acts as an Inducer IPTG or Isopropylthiogalactopyranoside

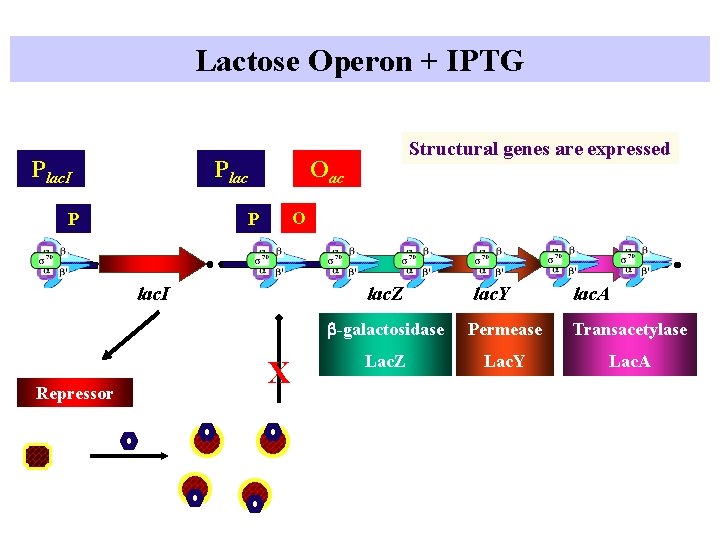
Lactose Operon + IPTG Plac. I Plac P Oac O P lac. I Repressor Structural genes are expressed lac. Z X lac. Y lac. A b-galactosidase Permease Transacetylase Lac. Z Lac. Y Lac. A

Synthetic Substrate b-galactosidase lactose glucose + galactose 5 -bromo-4 -chloro-3 -indolyl- b-D-galactoside b-galactosidase hydrolyses X-gal forming an indigo-type blue-green precipitate. Thus, bacteria which are positive for this enzyme produce blue colonies when grown in the presence of X-gal.