Gene Mutations Gene Mutations O A mutation is







- Slides: 19

Gene Mutations

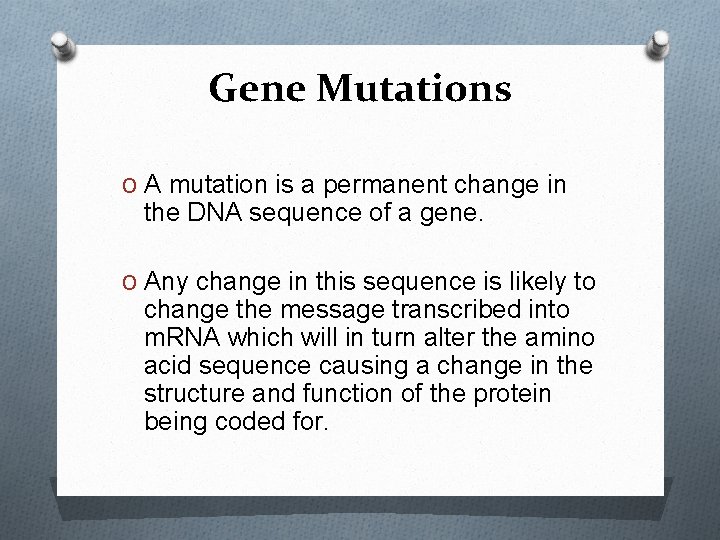
Gene Mutations O A mutation is a permanent change in the DNA sequence of a gene. O Any change in this sequence is likely to change the message transcribed into m. RNA which will in turn alter the amino acid sequence causing a change in the structure and function of the protein being coded for.

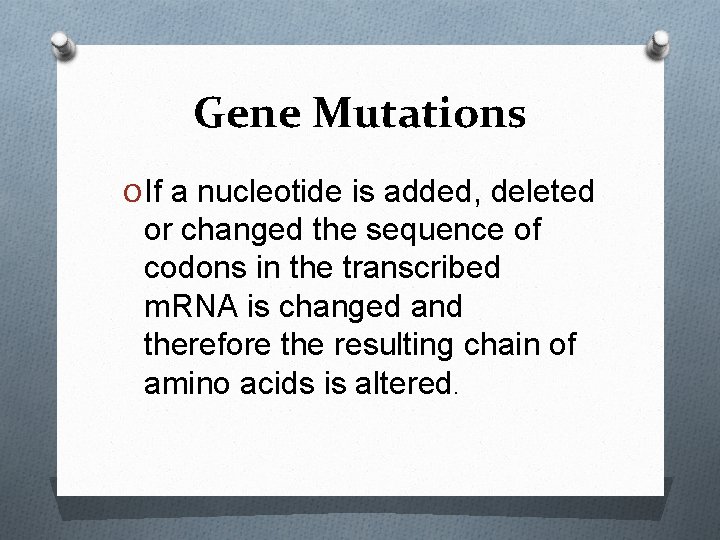
Gene Mutations O If a nucleotide is added, deleted or changed the sequence of codons in the transcribed m. RNA is changed and therefore the resulting chain of amino acids is altered.

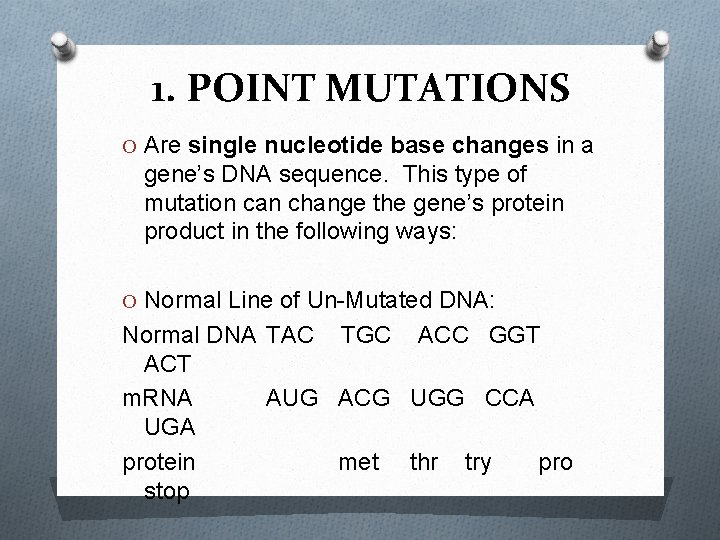
1. POINT MUTATIONS O Are single nucleotide base changes in a gene’s DNA sequence. This type of mutation can change the gene’s protein product in the following ways: O Normal Line of Un-Mutated DNA: Normal DNA TAC TGC ACC GGT ACT m. RNA AUG ACG UGG CCA UGA protein met thr try pro stop

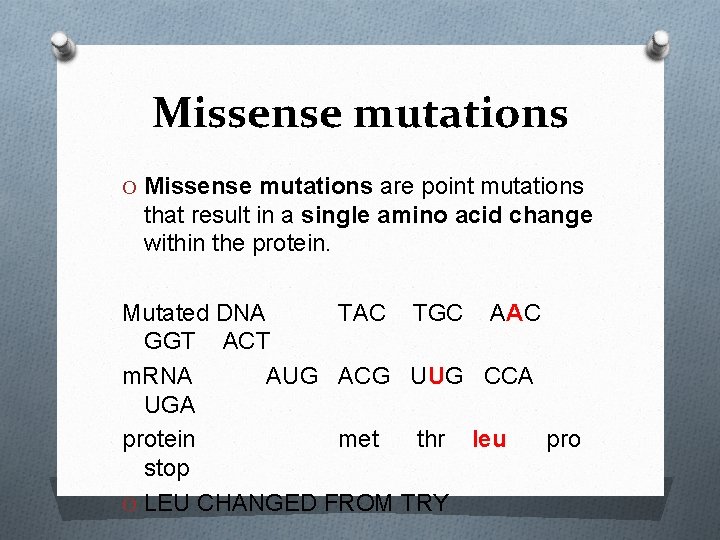
Missense mutations O Missense mutations are point mutations that result in a single amino acid change within the protein. Mutated DNA TAC TGC AAC GGT ACT m. RNA AUG ACG UUG CCA UGA protein met thr leu pro stop O LEU CHANGED FROM TRY

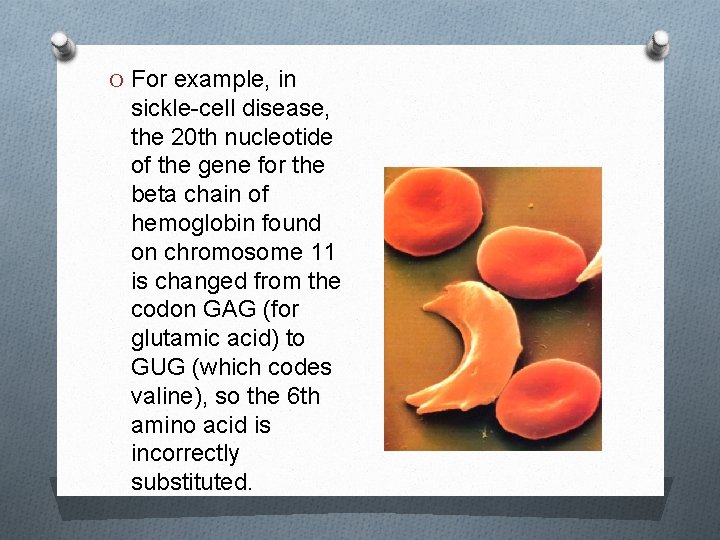
O For example, in sickle-cell disease, the 20 th nucleotide of the gene for the beta chain of hemoglobin found on chromosome 11 is changed from the codon GAG (for glutamic acid) to GUG (which codes valine), so the 6 th amino acid is incorrectly substituted.

Nonsense mutations O Nonsense mutations are point mutations that create a premature “translation stop signal” (or “stop” codon), causing the protein to be shortened. Mutated DNA TAC TGC ATC GGT ACT m. RNA AUG ACG UAG CCA UGA protein met thr STOP O STOP CHANGED FROM TRY

O Nonsense Mutations for cystic fibrosis have been found in almost 1000 combinations. Each of these mutations occurs in a huge gene that encodes a protein (of 1480 amino acids) called the cystic fibrosis transmembrane conductance regulator (CFTR). Unlike a missence mutation in sickle cell anemia, it can be various mutations that cause cystic fibrosis. O Ex: The substitution of a T for a C at nucleotide 1609 converted a glutamine codon (CAG) to a STOP codon (TAG). The protein produced by this patient had only the first 493 amino acids of the normal chain of 1480 and could not function.

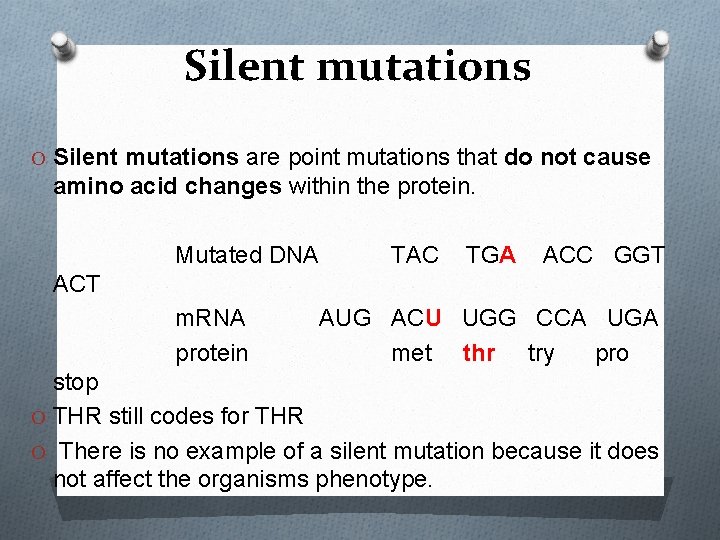
Silent mutations O Silent mutations are point mutations that do not cause amino acid changes within the protein. Mutated DNA TAC TGA ACC GGT ACT m. RNA protein AUG ACU UGG CCA UGA met thr try pro stop O THR still codes for THR O There is no example of a silent mutation because it does not affect the organisms phenotype.

2. FRAMESHIFT MUTATIONS O Result in a shift of the “reading frame” during protein translation. O There are two different types

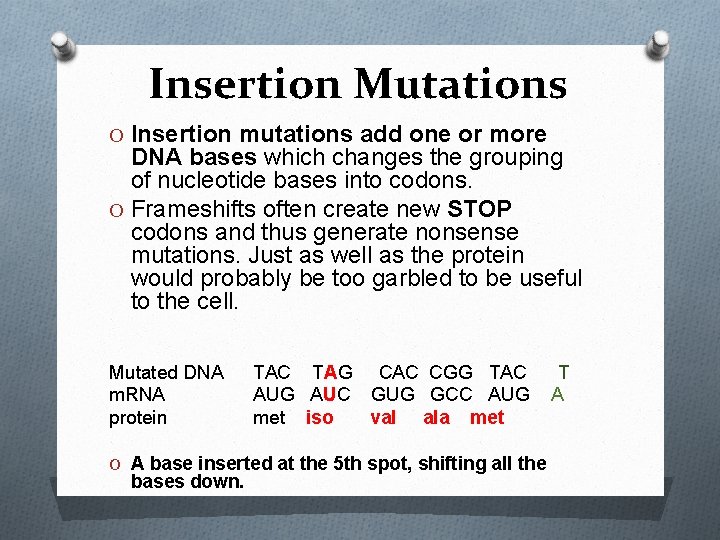
Insertion Mutations O Insertion mutations add one or more DNA bases which changes the grouping of nucleotide bases into codons. O Frameshifts often create new STOP codons and thus generate nonsense mutations. Just as well as the protein would probably be too garbled to be useful to the cell. Mutated DNA m. RNA protein TAC TAG CAC CGG TAC AUG AUC GUG GCC AUG met iso val ala met O A base inserted at the 5 th spot, shifting all the bases down. T A


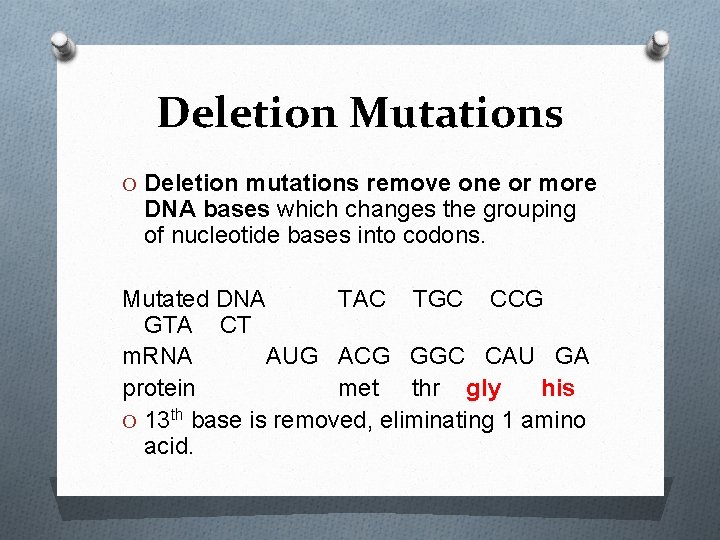
Deletion Mutations O Deletion mutations remove one or more DNA bases which changes the grouping of nucleotide bases into codons. Mutated DNA TAC TGC CCG GTA CT m. RNA AUG ACG GGC CAU GA protein met thr gly his O 13 th base is removed, eliminating 1 amino acid.

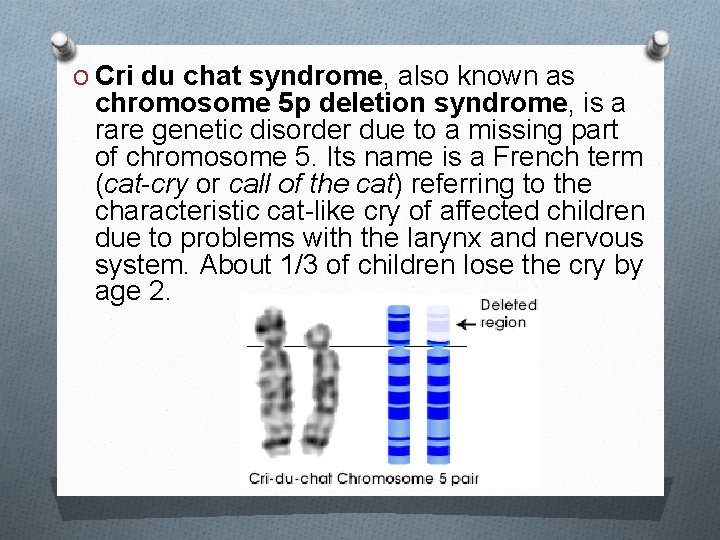
O Cri du chat syndrome, also known as chromosome 5 p deletion syndrome, is a rare genetic disorder due to a missing part of chromosome 5. Its name is a French term (cat-cry or call of the cat) referring to the characteristic cat-like cry of affected children due to problems with the larynx and nervous system. About 1/3 of children lose the cry by age 2.

O Other symptoms of cri du chat syndrome may include: O feeding problems because of difficulty swallowing and sucking. O low birth weight and poor growth. O severe cognitive, speech, and motor delays. O behavioral problems such as hyperactivity, aggression, tantrums, and repetitive movements. O unusual facial features which may change over time. O excessive drooling.

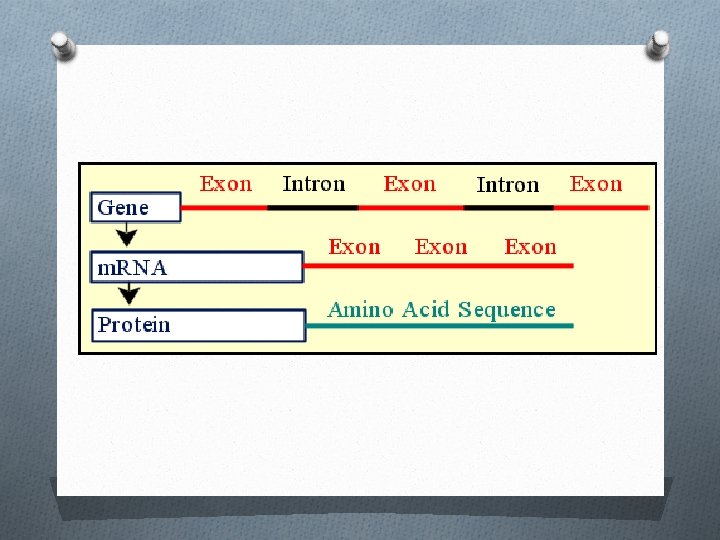
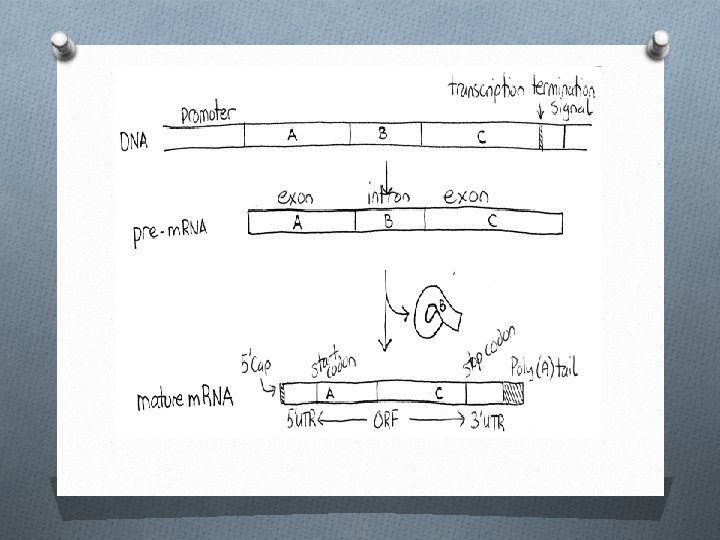
Introns and Exons O Exons – segments of DNA that code for amino acids that will become part of a protein. O Exons are not found next to each other. They are separated by sections of DNA called introns.

O Introns – segments of DNA that do not code for amino acids of a protein. O When a gene is transcribed, both the introns and exons are made into RNA. O The introns must then be cut out and the remaining exon pieces must be joined together. O Before this happens, two other strings of nucleotides are added to the ends of the RNA. One is called a cap and the other is called a tail.

