Gene Finding Finding Genes Prokaryotes Genome under 10
















- Slides: 26

Gene Finding

Finding Genes • Prokaryotes – Genome under 10 Mb – >85% of sequence codes for proteins • Eukaryotes – Large Genomes (up to 10 Gb) – 1 -3% coding for vertebrates

Introns • Humans – – 95% of genes have introns 10% of genes have more than 20 introns Some have more than 60 Largest Gene (Duchenne muscular dystrophy locus) spans >2 Mb (more than a prokaryote) – Average exon = 150 b – Introns can interrupt Open Reading Frame at any position, even within a codon – ORF finding is not sufficient for Eukaryotic genomes

Open Reading Frames in Bacteria • Without introns, look for long open reading frame (start codon ATG, … , stop codon TAA, TAG, TGA) • Short genes are missed (<300 nucleotides) • Shadow genes (overlapping open reading frames on opposite DNA strands) are hard to detect • Some genes start with UUG, AUA, UUA and CUG for start codon • Some genes use TGA to create selenocysteine and it is not a stop codon

Eukaryotes • Maps are used as scaffolding during sequencing • Recombination is used to predict the distance genes are from each other (the further apart two loci are on the chromosome, the more likely they are to be separated by recombination during meiosis) • Pedigree analysis

Gene Finding in Eukaryotes • Look for strongly conserved regions • RNA blots - map expressed RNA to DNA • Identification of CPG islands – Short stretches of CG rich DNA are associated with the promoters of vertebrate genes • Exon Trapping - put questionable clone between two exons that are expressed. If there is a gene, it will be spliced into the mature transcript

Computational methods • Signals - TATA box and other sequences – TATA box is found 30 bp upstream from about 70% of the genes • Content - Coding DNA and non-coding DNA differ in terms of Hexamer frequency (frequency with which specific 6 nucleotide strings are used) – Some organisms prefer different codons for the same amino acid • Homology - blast for sequence in other organisms

Genome Browser • http: //genome. ucsc. edu/ • Tables • Genome browser

Non-coding RNA genes • Ribosomal r. RNA, transfer t. RNA can be recognized by stochastic context-free grammars • Detection is still an open problem




Hidden Markov Models (HMMs) • Provide a probabilistic view of a process that we don’t fully understand • The model can be trained with data we don’t understand to learn patterns • You get to implement one for the first lab!!

State Transitions Markov Model Example. --x = States of the Markov model -- a = Transition probabilities -- b = Output probabilities -- y = Observable outputs -How does this differ from a Finite State machine? -Why is it a Markov process?

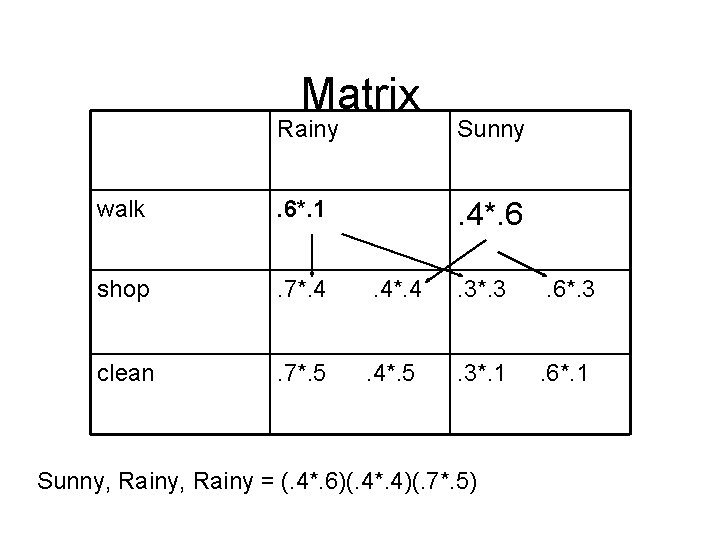
Example • Distant friend that you talk to daily about his activities (walk, shop, clean) • You believe that the weather is a discrete Markov chain (no memory) with two states (rainy, sunny), but you cant observe them directly. You know the average weather patterns

Formal Description states = ('Rainy', 'Sunny') observations = ('walk', 'shop', 'clean') start_probability = {'Rainy': 0. 6, 'Sunny': 0. 4} transition_probability = { 'Rainy' : {'Rainy': 0. 7, 'Sunny': 0. 3}, 'Sunny' : {'Rainy': 0. 4, 'Sunny': 0. 6}, } emission_probability = { 'Rainy' : {'walk': 0. 1, 'shop': 0. 4, 'clean': 0. 5}, 'Sunny' : {'walk': 0. 6, 'shop': 0. 3, 'clean': 0. 1}, }

Observations • Given (walk, shop, clean) – What is the probability of this sequence of observations? (is he really still at home, or did he skip the country) – What was the most likely sequence of rainy/sunny days?

Matrix Rainy Sunny walk . 6*. 1 . 4*. 6 shop . 7*. 4 . 4*. 4 . 3*. 3 . 6*. 3 clean . 7*. 5 . 4*. 5 . 3*. 1 . 6*. 1 Sunny, Rainy = (. 4*. 6)(. 4*. 4)(. 7*. 5)

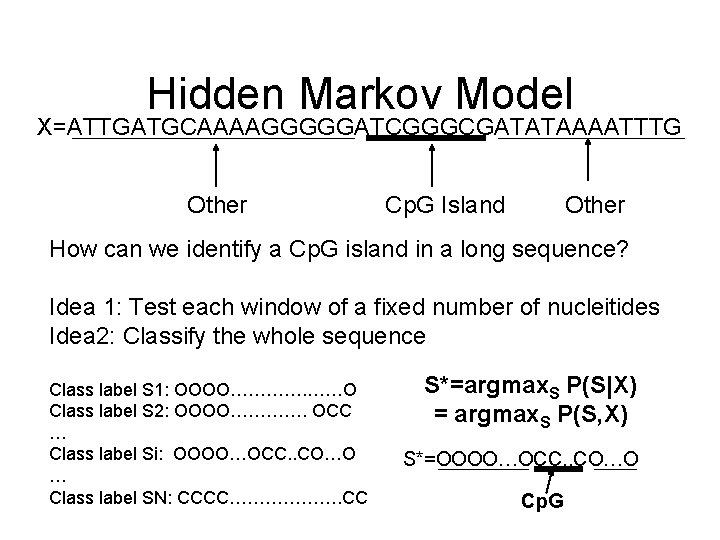
The Cp. G island problem • Methylation in human genome – “CG” -> “TG” happens in most places except “start regions” of genes and within genes – Cp. G islands = 100 -1, 000 bases before a gene starts • Question – Given a long sequence, how would we find the Cp. G islands in it?

Hidden Markov Model X=ATTGATGCAAAAGGGGGATCGGGCGATATAAAATTTG Other Cp. G Island Other How can we identify a Cp. G island in a long sequence? Idea 1: Test each window of a fixed number of nucleitides Idea 2: Classify the whole sequence Class label S 1: OOOO…………. ……O Class label S 2: OOOO…………. OCC … Class label Si: OOOO…OCC. . CO…O … Class label SN: CCCC………………. CC S*=argmax. S P(S|X) = argmax. S P(S, X) S*=OOOO…OCC. . CO…O Cp. G

HMM is just one way of modeling p(X, S)…

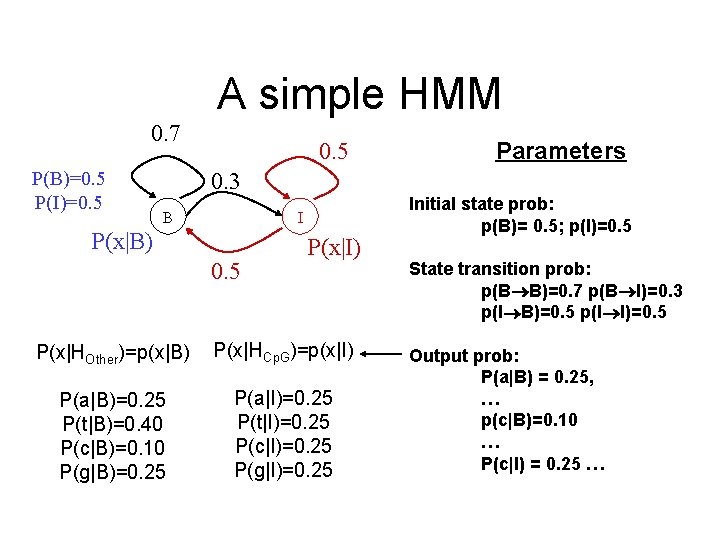
A simple HMM 0. 7 P(B)=0. 5 P(I)=0. 5 Parameters 0. 3 B I P(x|B) 0. 5 P(x|I) P(x|HOther)=p(x|B) P(x|HCp. G)=p(x|I) P(a|B)=0. 25 P(t|B)=0. 40 P(c|B)=0. 10 P(g|B)=0. 25 P(a|I)=0. 25 P(t|I)=0. 25 P(c|I)=0. 25 P(g|I)=0. 25 Initial state prob: p(B)= 0. 5; p(I)=0. 5 State transition prob: p(B B)=0. 7 p(B I)=0. 3 p(I B)=0. 5 p(I I)=0. 5 Output prob: P(a|B) = 0. 25, … p(c|B)=0. 10 … P(c|I) = 0. 25 …

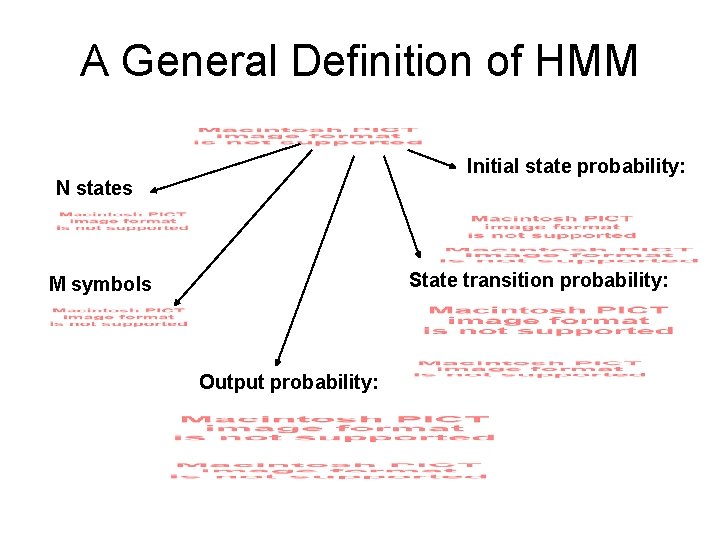
A General Definition of HMM Initial state probability: N states State transition probability: M symbols Output probability:

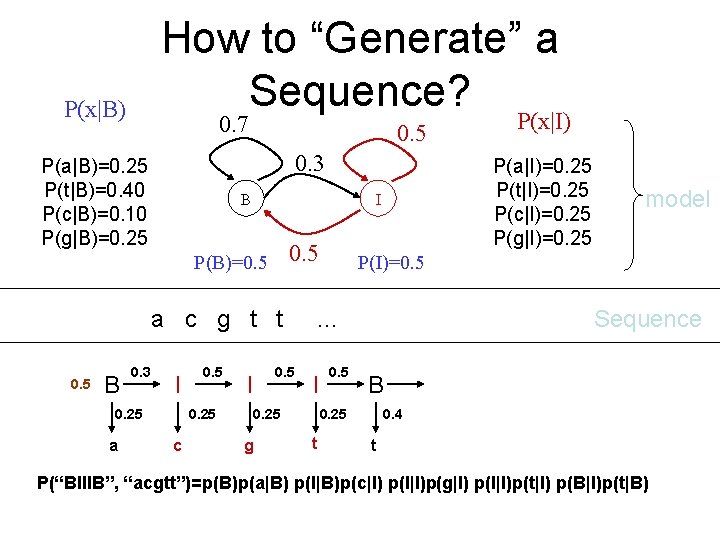
P(x|B) How to “Generate” a Sequence? 0. 7 P(x|I) 0. 5 0. 3 P(a|B)=0. 25 P(t|B)=0. 40 P(c|B)=0. 10 P(g|B)=0. 25 B P(B)=0. 5 a c g t B I I I B P(a|I)=0. 25 P(t|I)=0. 25 P(c|I)=0. 25 P(g|I)=0. 25 I 0. 5 t P(I)=0. 5 … B model Sequence B I B states B I I B …… Given a model, follow a path to generate the observations.

How to “Generate” a Sequence? P(x|B) 0. 7 0. 5 0. 3 P(a|B)=0. 25 P(t|B)=0. 40 P(c|B)=0. 10 P(g|B)=0. 25 B 0. 5 a c g t t 0. 5 B 0. 3 I 0. 25 a 0. 5 0. 25 c P(a|I)=0. 25 P(t|I)=0. 25 P(c|I)=0. 25 P(g|I)=0. 25 I P(B)=0. 5 I 0. 25 g 0. 5 Sequence B 0. 25 t model P(I)=0. 5 … I P(x|I) 0. 4 t P(“BIIIB”, “acgtt”)=p(B)p(a|B) p(I|B)p(c|I) p(I|I)p(g|I) p(I|I)p(t|I) p(B|I)p(t|B)

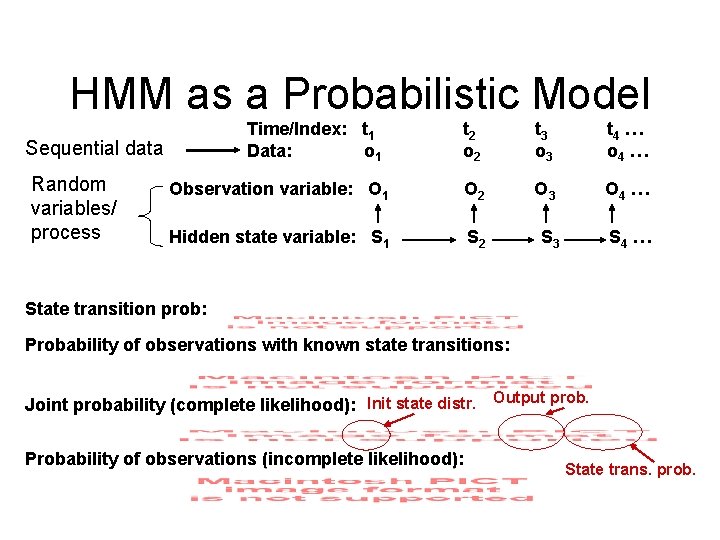
HMM as a Probabilistic Model Time/Index: t 1 Data: o 1 t 2 o 2 t 3 o 3 t 4 … o 4 … Observation variable: O 1 O 2 O 3 O 4 … Hidden state variable: S 1 S 2 S 3 S 4 … Sequential data Random variables/ process State transition prob: Probability of observations with known state transitions: Joint probability (complete likelihood): Init state distr. Output prob. Probability of observations (incomplete likelihood): State trans. prob.