Gene Expression and Signaling Pathways in Yeast Classical




- Slides: 33

Gene Expression and Signaling Pathways in Yeast .

Classical Genetics Genotype: The genetic makeup of an organism Phenotype: The observed “behavior” Basic Assumption: Genotype phenotype

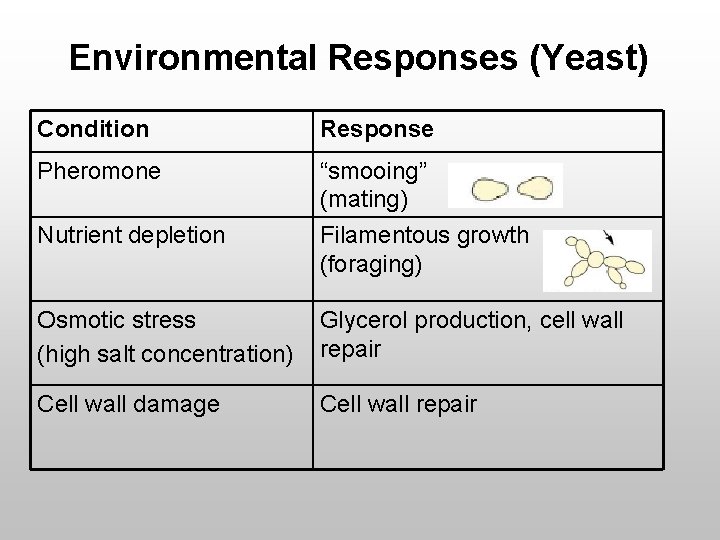
Environmental Responses (Yeast) Condition Response Pheromone “smooing” (mating) Nutrient depletion Filamentous growth (foraging) Osmotic stress (high salt concentration) Glycerol production, cell wall repair Cell wall damage Cell wall repair

Genetic Screens u Search for genes whose removal (KO) disrupt desired response u Classical genetic screens identified many genes that are involved in yeast response to environmental cues. Can we understand these processes better?


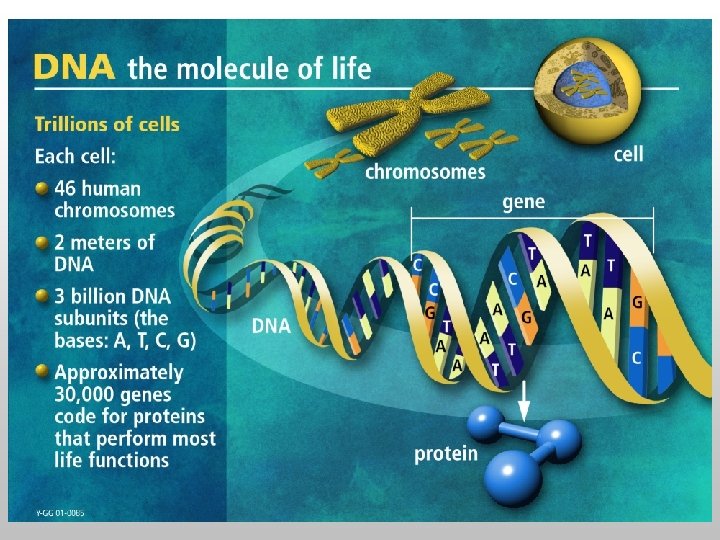
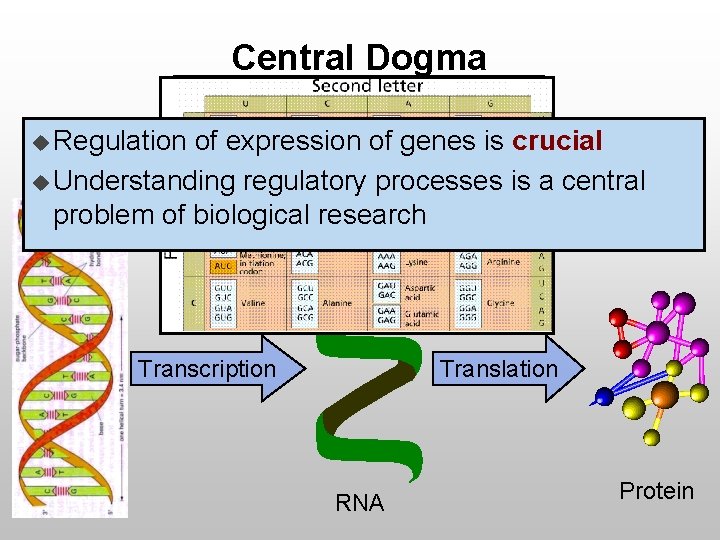
Central Dogma u Regulation of expression of genes is crucial u Understanding regulatory processes is a central problem of biological research Transcription Translation RNA Protein

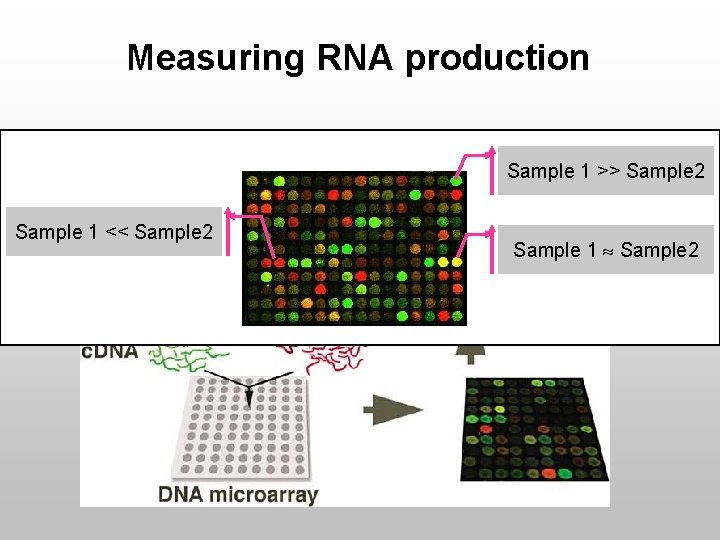
Measuring RNA production Sample 1 >> Sample 2 Sample 1 << Sample 2 Sample 1 Sample 2


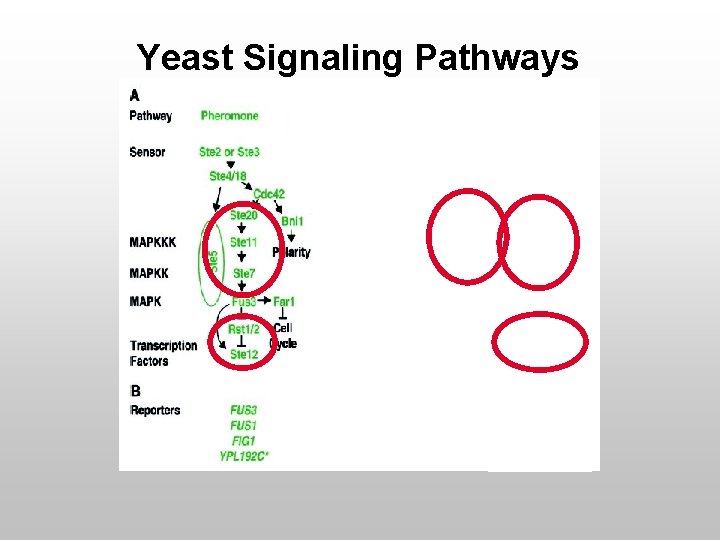
Yeast Signaling Pathways

Goals u Comprehensive map of genes involved in different responses u Understanding the function of different components in the pathway Meta-goal: u Utility of gene expression for studying signaling pathways

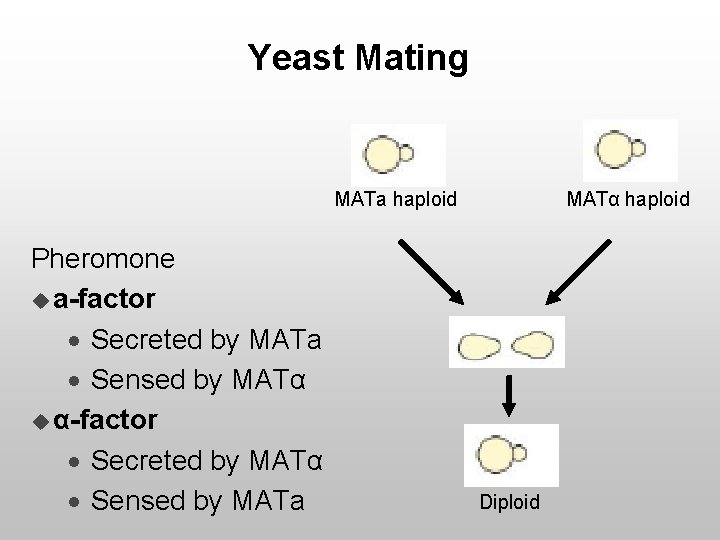
Yeast Mating MATa haploid Pheromone u a-factor · Secreted by MATa · Sensed by MATα u α-factor · Secreted by MATα · Sensed by MATa MATα haploid Diploid

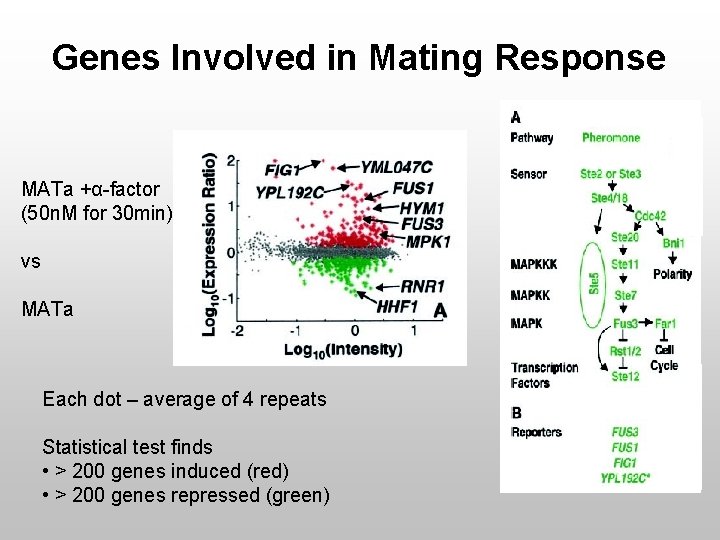
Genes Involved in Mating Response MATa +α-factor (50 n. M for 30 min) vs MATa Each dot – average of 4 repeats Statistical test finds • > 200 genes induced (red) • > 200 genes repressed (green)

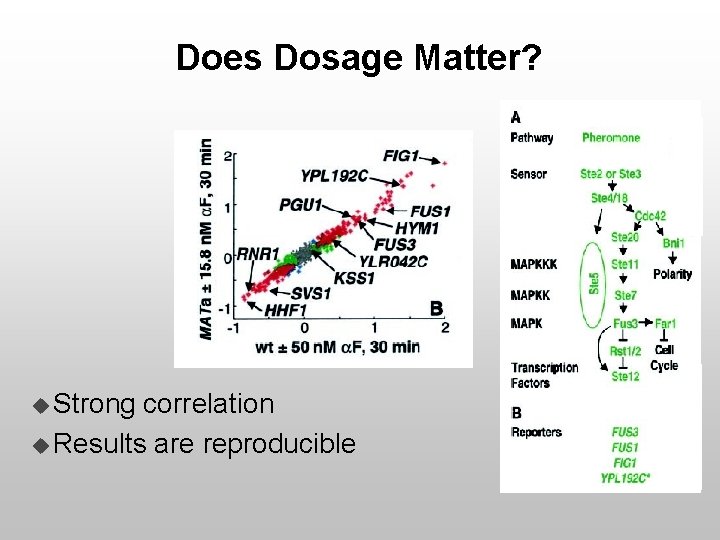
Does Dosage Matter? u Strong correlation u Results are reproducible

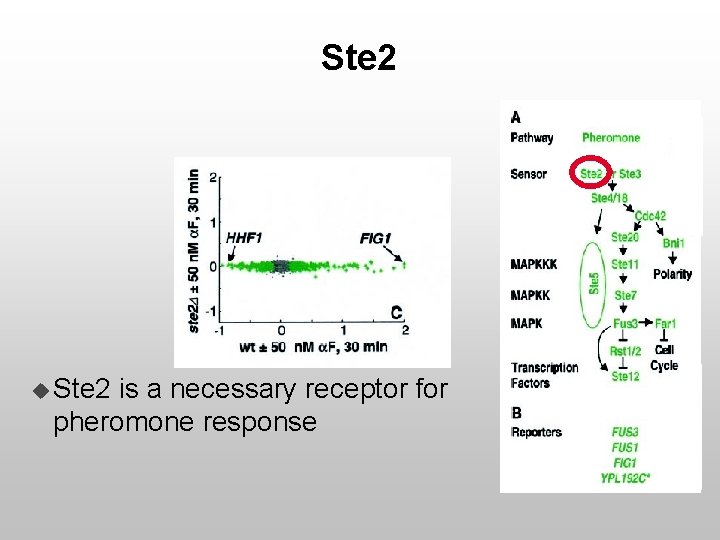
Ste 2 u Ste 2 is a necessary receptor for pheromone response

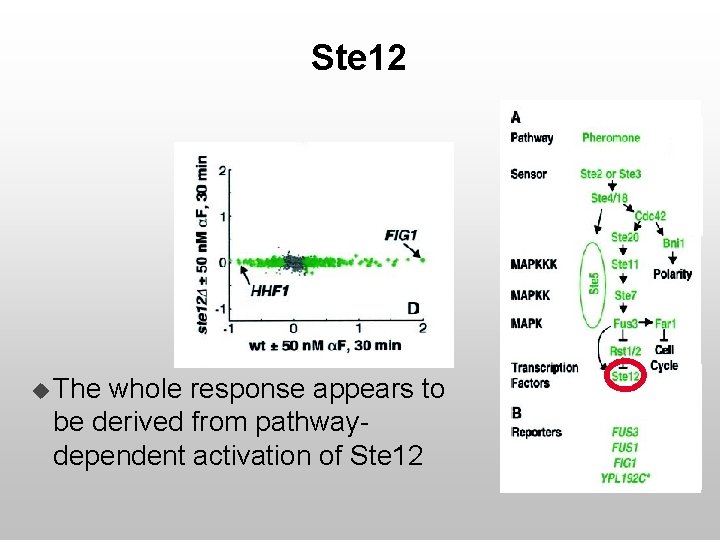
Ste 12 u The whole response appears to be derived from pathwaydependent activation of Ste 12

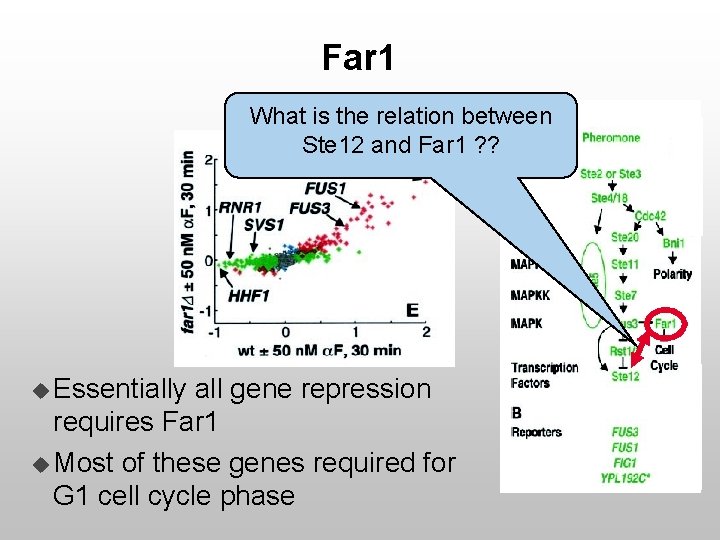
Far 1 What is the relation between Ste 12 and Far 1 ? ? u Essentially all gene repression requires Far 1 u Most of these genes required for G 1 cell cycle phase

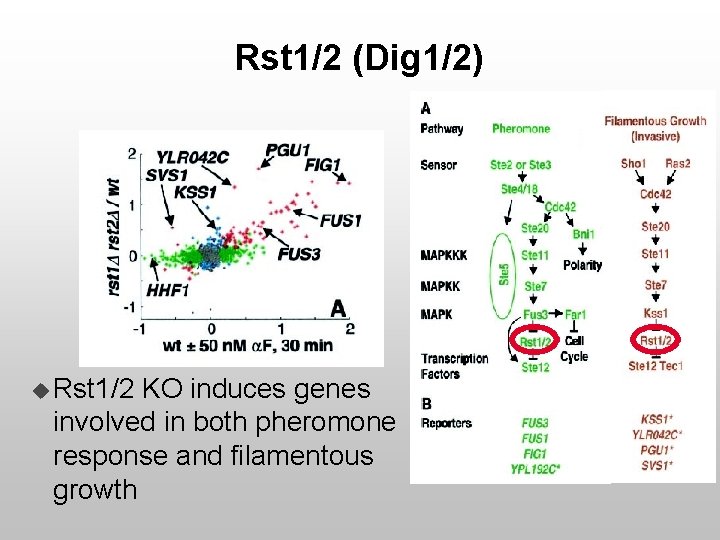
Rst 1/2 (Dig 1/2) u Rst 1/2 KO induces genes involved in both pheromone response and filamentous growth

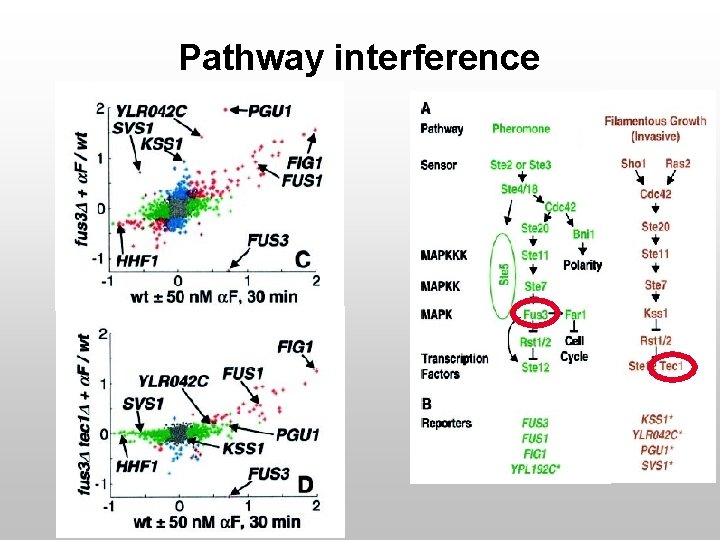
Pathway interference

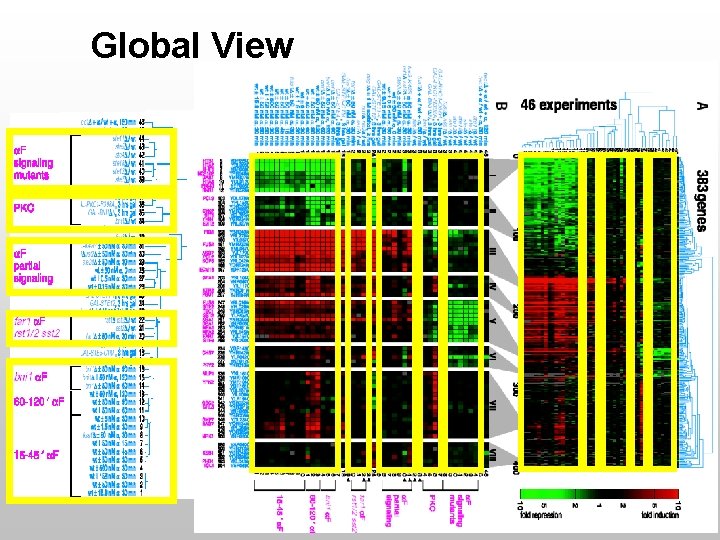
Global View

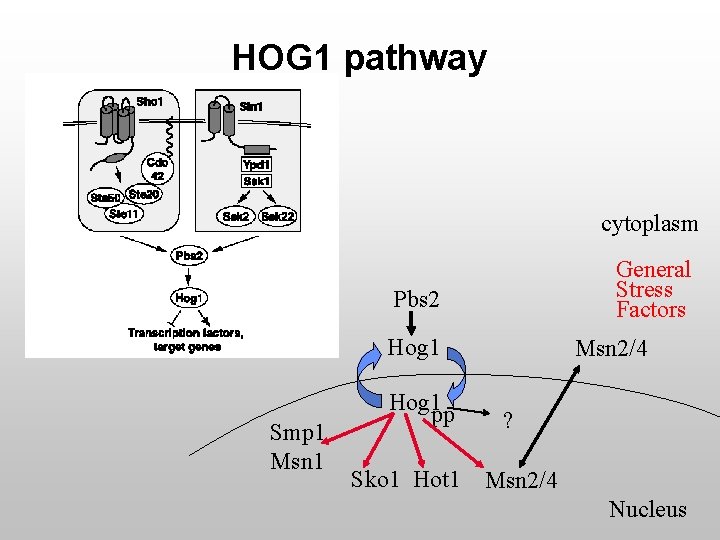
Goals u Closer look at HOG 1 pathway · Activity of HOG 1 · Roles of components upstream of HOG 1

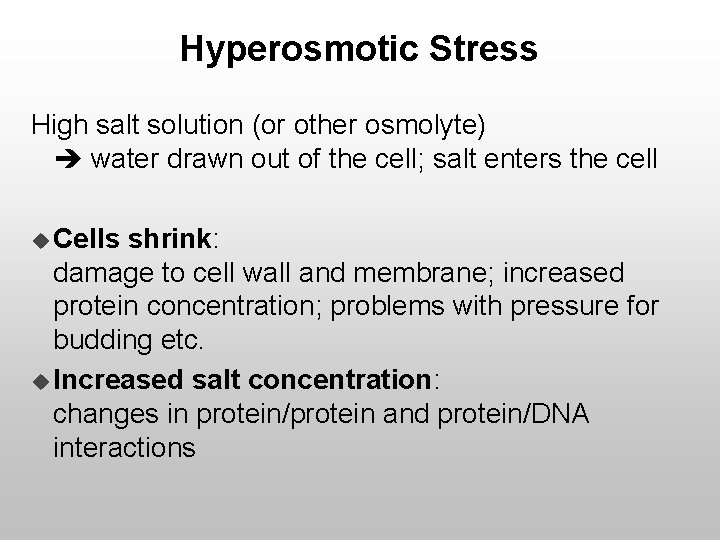
Hyperosmotic Stress High salt solution (or other osmolyte) water drawn out of the cell; salt enters the cell u Cells shrink: damage to cell wall and membrane; increased protein concentration; problems with pressure for budding etc. u Increased salt concentration: changes in protein/protein and protein/DNA interactions

Hyperosmotic Stress Response u Production of an osmolyte: · Glycerol is synthesized to balance osmolarity inside/outside · Change in metabolism to accommodate this u Removal of salt: · Upregulation/downregulation of appropriate transporters for Na. Cl, KCl and other ions u Dealing with stress: · Expression of chaperones and other general stress response genes · Shutting down non-essential stuff

HOG 1 pathway cytoplasm General Stress Factors Pbs 2 Hog 1 Smp 1 Msn 1 Hog 1 pp Sko 1 Hot 1 Msn 2/4 ? Msn 2/4 Nucleus

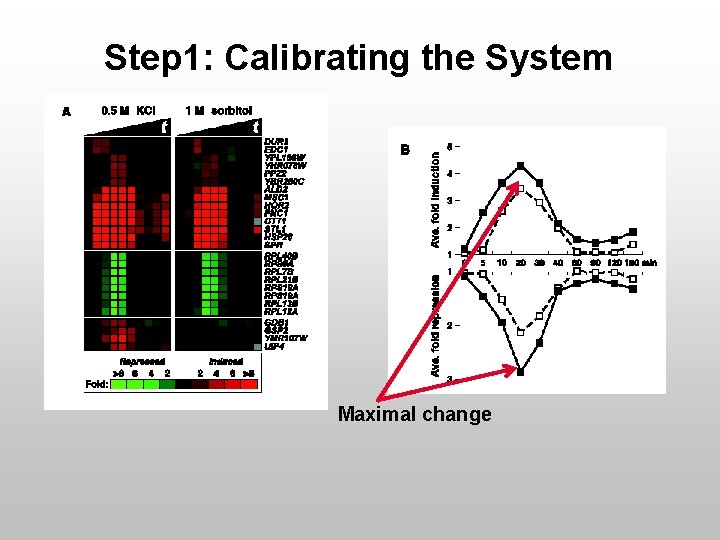
Step 1: Calibrating the System Maximal change

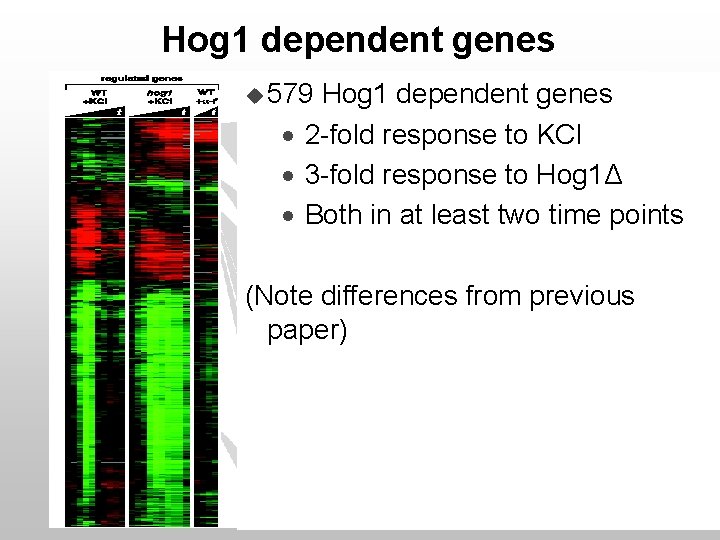
Hog 1 dependent genes u 579 Hog 1 dependent genes · 2 -fold response to KCl · 3 -fold response to Hog 1Δ · Both in at least two time points (Note differences from previous paper)

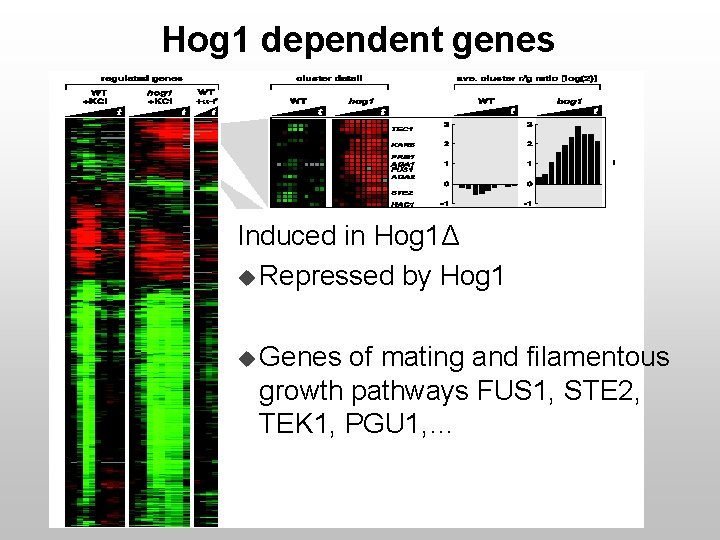
Hog 1 dependent genes Induced in Hog 1Δ u Repressed by Hog 1 u Genes of mating and filamentous growth pathways FUS 1, STE 2, TEK 1, PGU 1, …

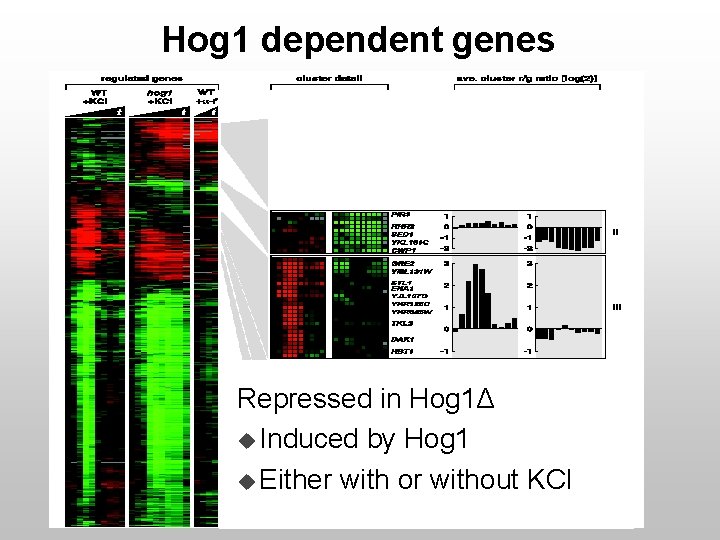
Hog 1 dependent genes Repressed in Hog 1Δ u Induced by Hog 1 u Either with or without KCl

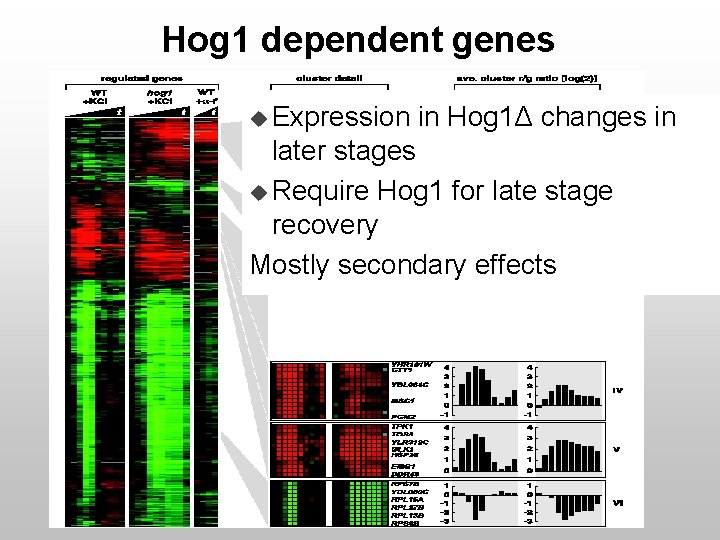
Hog 1 dependent genes u Expression in Hog 1Δ changes in later stages u Require Hog 1 for late stage recovery Mostly secondary effects

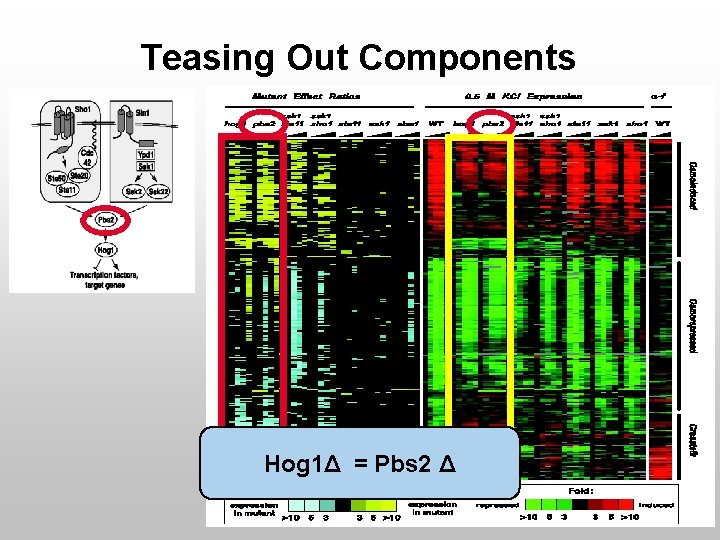
Teasing Out Components Hog 1Δ = Pbs 2 Δ

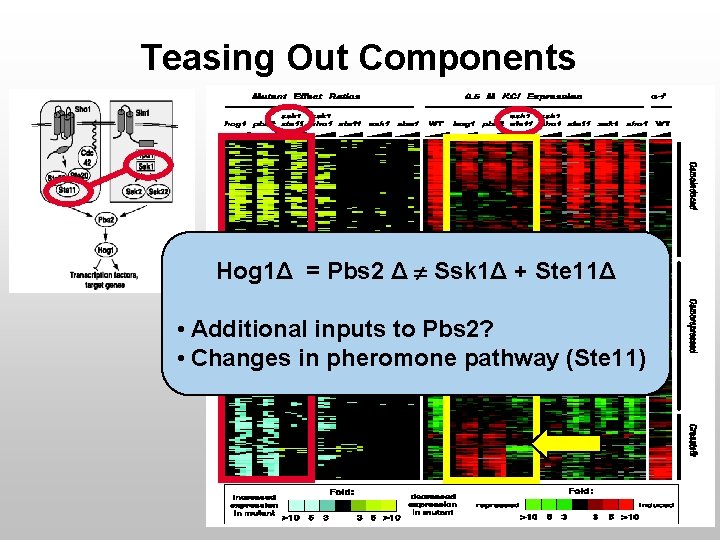
Teasing Out Components Hog 1Δ = Pbs 2 Δ Ssk 1Δ + Ste 11Δ • Additional inputs to Pbs 2? • Changes in pheromone pathway (Ste 11)

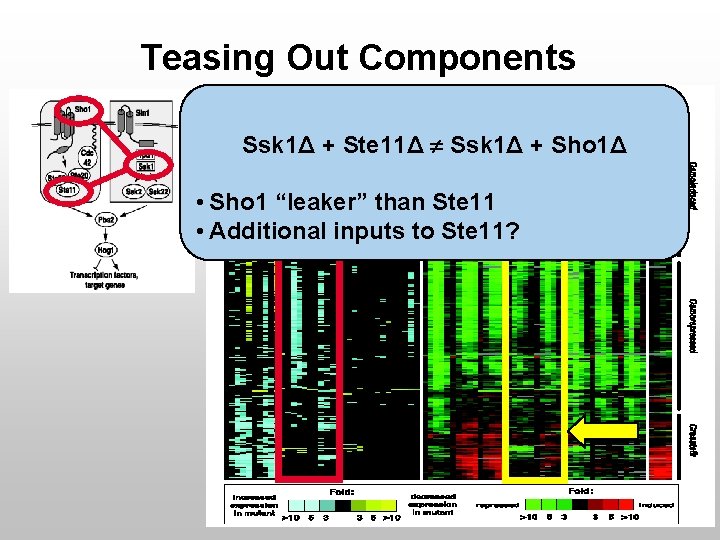
Teasing Out Components Ssk 1Δ + Ste 11Δ Ssk 1Δ + Sho 1Δ • Sho 1 “leaker” than Ste 11 • Additional inputs to Ste 11?

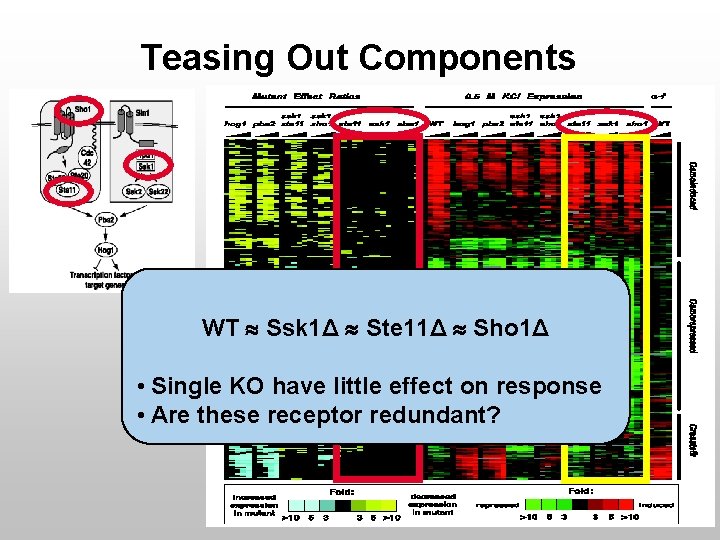
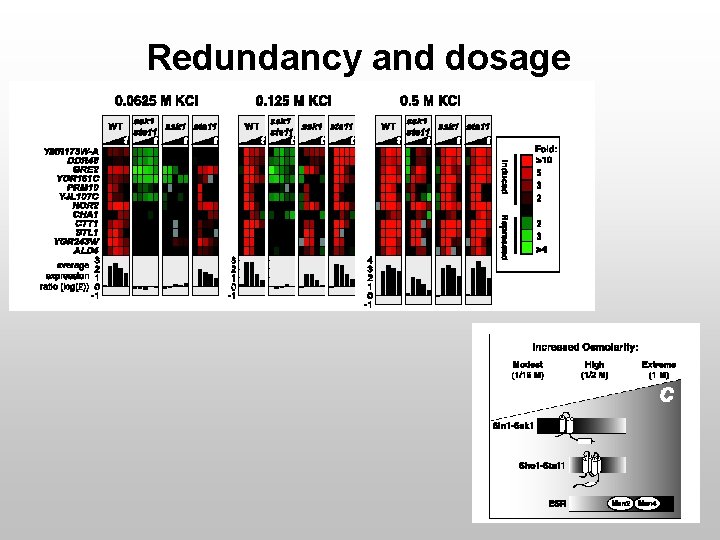
Teasing Out Components WT Ssk 1Δ Ste 11Δ Sho 1Δ • Single KO have little effect on response • Are these receptor redundant?

Redundancy and dosage