Gene Editing By Marwa Moustafa What is Gene

Gene Editing By / Marwa Moustafa

What is Gene Editing?
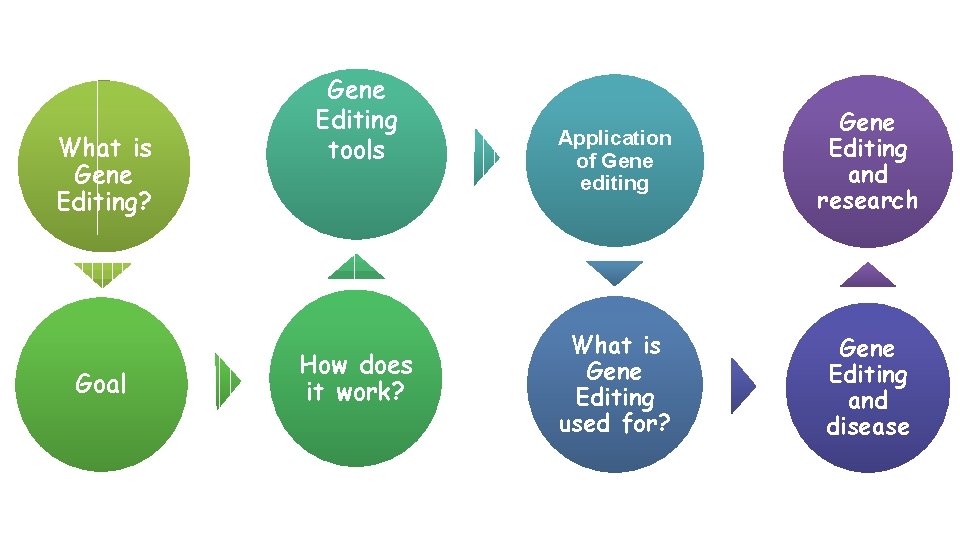
What is Gene Editing? Goal Gene Editing tools How does it work? Application of Gene editing Gene Editing and research What is Gene Editing used for? Gene Editing and disease
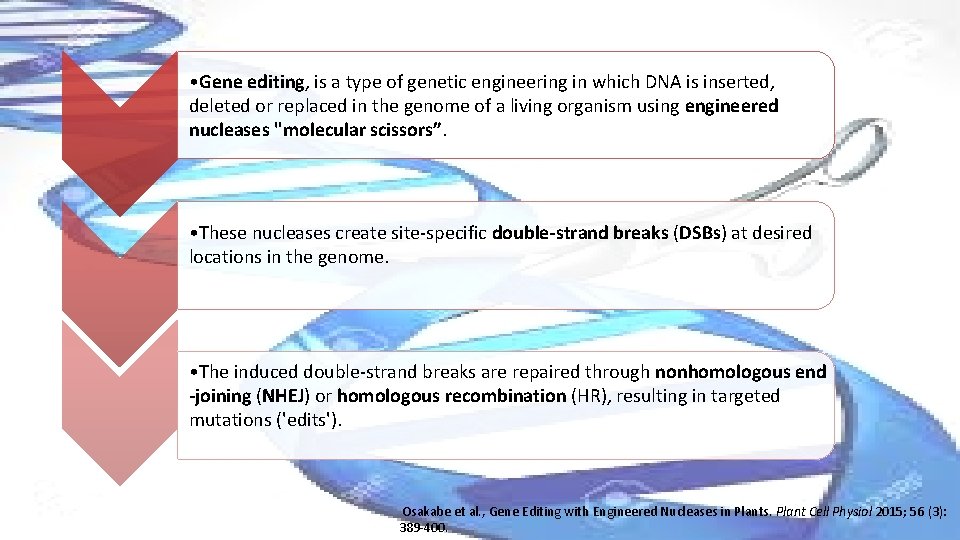
• Gene editing, is a type of genetic engineering in which DNA is inserted, deleted or replaced in the genome of a living organism using engineered nucleases "molecular scissors”. • These nucleases create site-specific double-strand breaks (DSBs) at desired locations in the genome. • The induced double-strand breaks are repaired through nonhomologous end -joining (NHEJ) or homologous recombination (HR), resulting in targeted mutations ('edits'). Osakabe et al. , Gene Editing with Engineered Nucleases in Plants. Plant Cell Physiol 2015; 56 (3): 389 -400.
The ultimate maneuver in Gene Editing is to replace a defective gene with a normal allele at its natural chromosomal location Carroll, D. (2008). Zinc-finger Nucleases as Gene Therapy Agents. Gene Therapy, 15(22), 1463– 1468.
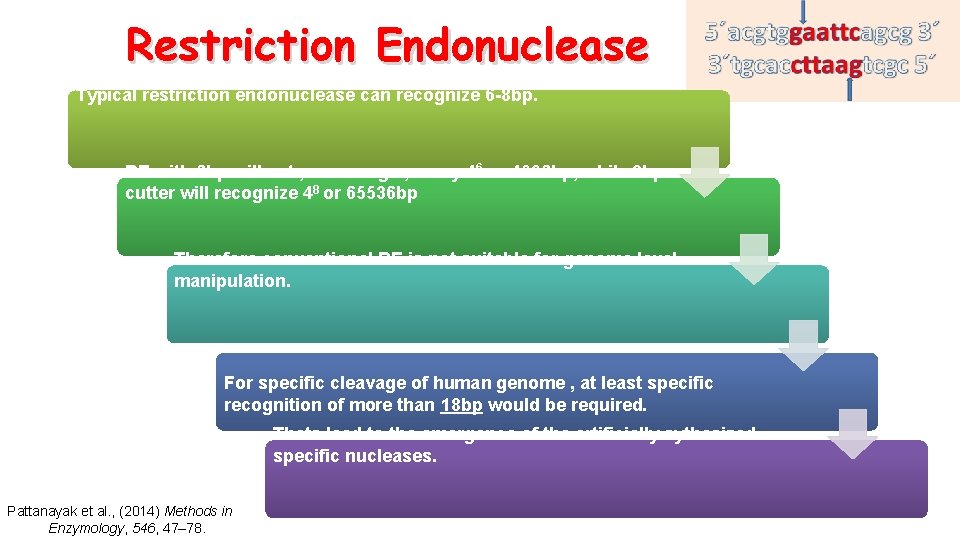
Restriction Endonuclease Typical restriction endonuclease can recognize 6 -8 bp. RE with 6 bp will cut , on average , every 46 or 4096 bp, while 8 bp cutter will recognize 48 or 65536 bp Therefore conventional RE is not suitable for genome level manipulation. For specific cleavage of human genome , at least specific recognition of more than 18 bp would be required. Thats lead to the emergence of the artificially sythesized specific nucleases. Pattanayak et al. , (2014) Methods in Enzymology, 546, 47– 78.

What are the DNA Editing mechanisms?
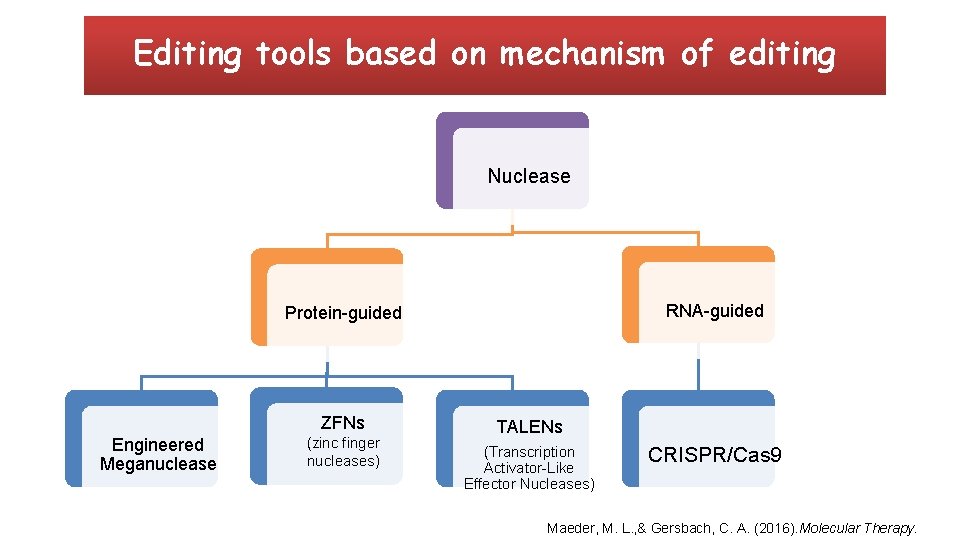
Editing tools based on mechanism of editing Nuclease RNA-guided Protein-guided ZFNs Engineered Meganuclease (zinc finger nucleases) TALENs (Transcription Activator-Like Effector Nucleases) CRISPR/Cas 9 Maeder, M. L. , & Gersbach, C. A. (2016). Molecular Therapy.
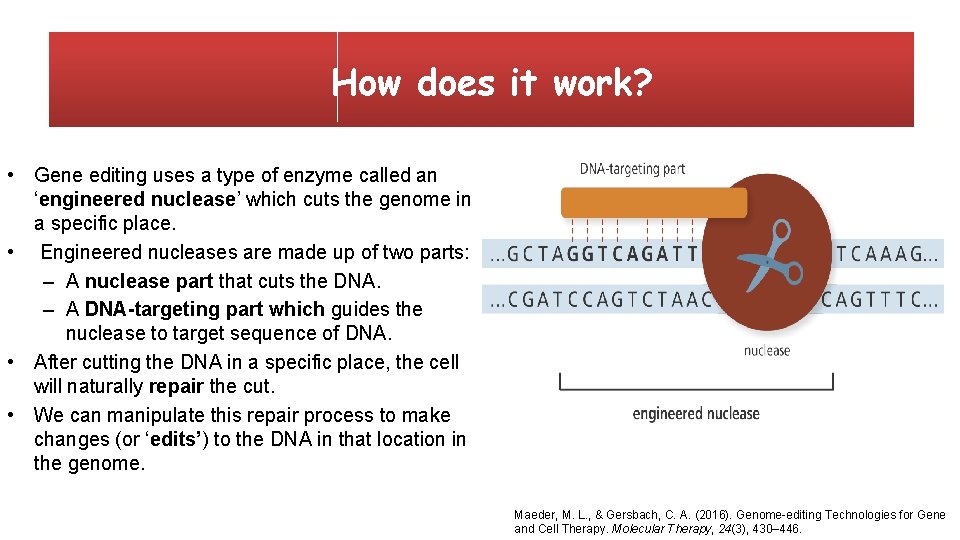
How does it work? • Gene editing uses a type of enzyme called an ‘engineered nuclease’ which cuts the genome in a specific place. • Engineered nucleases are made up of two parts: – A nuclease part that cuts the DNA. – A DNA-targeting part which guides the nuclease to target sequence of DNA. • After cutting the DNA in a specific place, the cell will naturally repair the cut. • We can manipulate this repair process to make changes (or ‘edits’) to the DNA in that location in the genome. Maeder, M. L. , & Gersbach, C. A. (2016). Genome-editing Technologies for Gene and Cell Therapy. Molecular Therapy, 24(3), 430– 446.
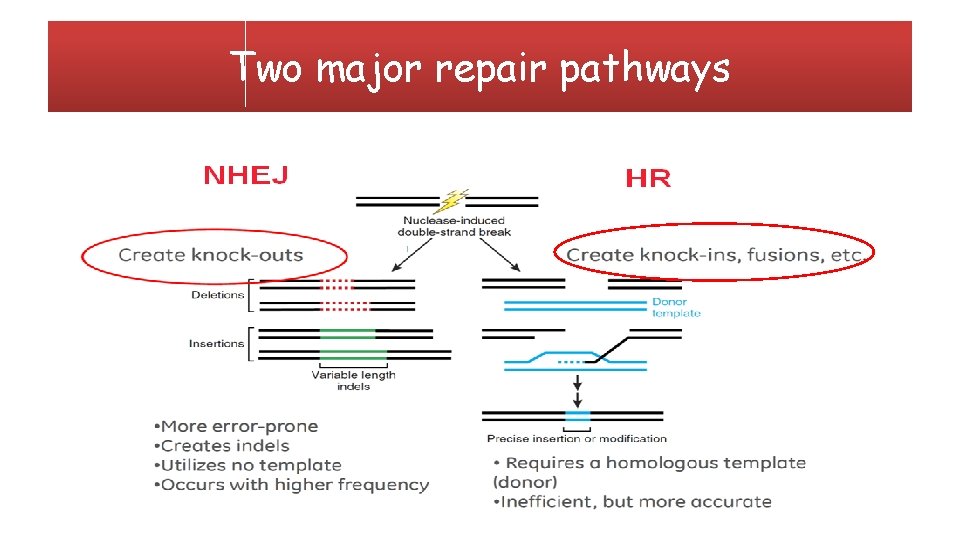
Two major repair pathways
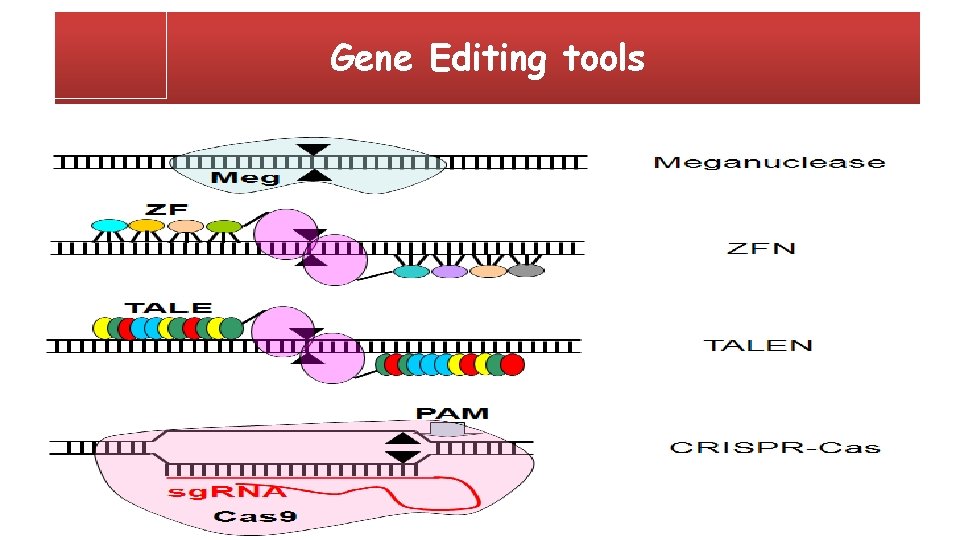
Gene Editing tools
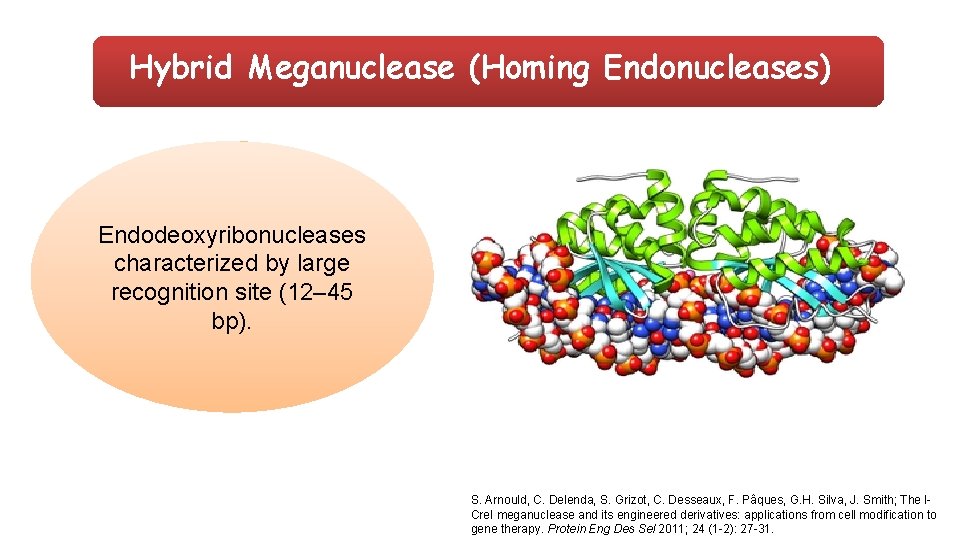
Hybrid Meganuclease (Homing Endonucleases) Endodeoxyribonucleases characterized by large recognition site (12– 45 bp). S. Arnould, C. Delenda, S. Grizot, C. Desseaux, F. Pâques, G. H. Silva, J. Smith; The ICre. I meganuclease and its engineered derivatives: applications from cell modification to gene therapy. Protein Eng Des Sel 2011; 24 (1 -2): 27 -31.
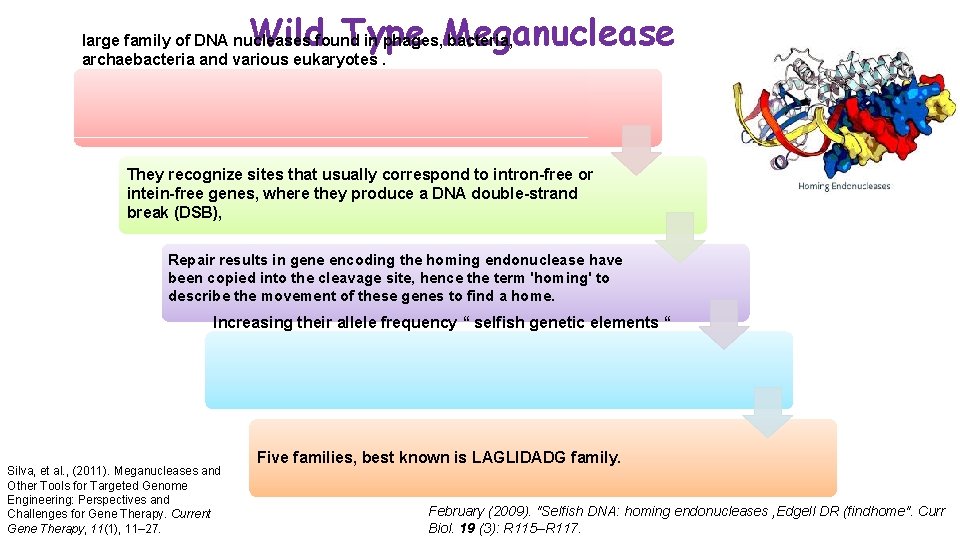
Wild Type Meganuclease large family of DNA nucleases found in phages, bacteria, archaebacteria and various eukaryotes. They recognize sites that usually correspond to intron-free or intein-free genes, where they produce a DNA double-strand break (DSB), Repair results in gene encoding the homing endonuclease have been copied into the cleavage site, hence the term 'homing' to describe the movement of these genes to find a home. Increasing their allele frequency “ selfish genetic elements “ Silva, et al. , (2011). Meganucleases and Other Tools for Targeted Genome Engineering: Perspectives and Challenges for Gene Therapy. Current Gene Therapy, 11(1), 11– 27. Five families, best known is LAGLIDADG family. February (2009). "Selfish DNA: homing endonucleases , Edgell DR (findhome". Curr Biol. 19 (3): R 115–R 117.
Engineered Meganuclease To create a tailor-made Meganucleases • Modify the specificity of existing meganucleases • The possibility of associating or fusing protein domains from different enzymes. Limitations • potential genotoxicity • lower efficacy. Huirong et al. , (2010). "Heritable targeted mutagenesis in maize using a designed endonuclease". The Plant Journal. 61 (1): 176– 87.
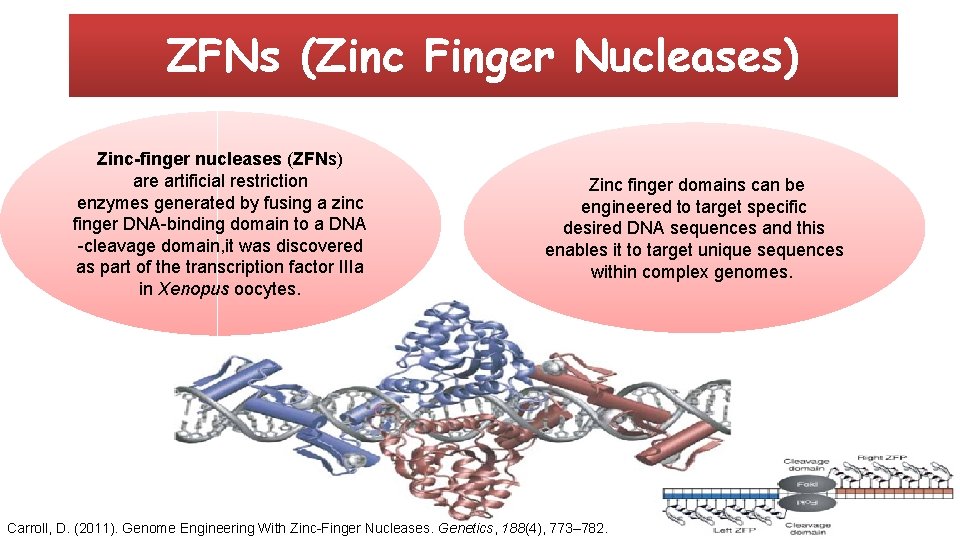
ZFNs (Zinc Finger Nucleases) Zinc-finger nucleases (ZFNs) are artificial restriction enzymes generated by fusing a zinc finger DNA-binding domain to a DNA -cleavage domain, it was discovered as part of the transcription factor IIIa in Xenopus oocytes. Zinc finger domains can be engineered to target specific desired DNA sequences and this enables it to target unique sequences within complex genomes. Carroll, D. (2011). Genome Engineering With Zinc-Finger Nucleases. Genetics, 188(4), 773– 782.
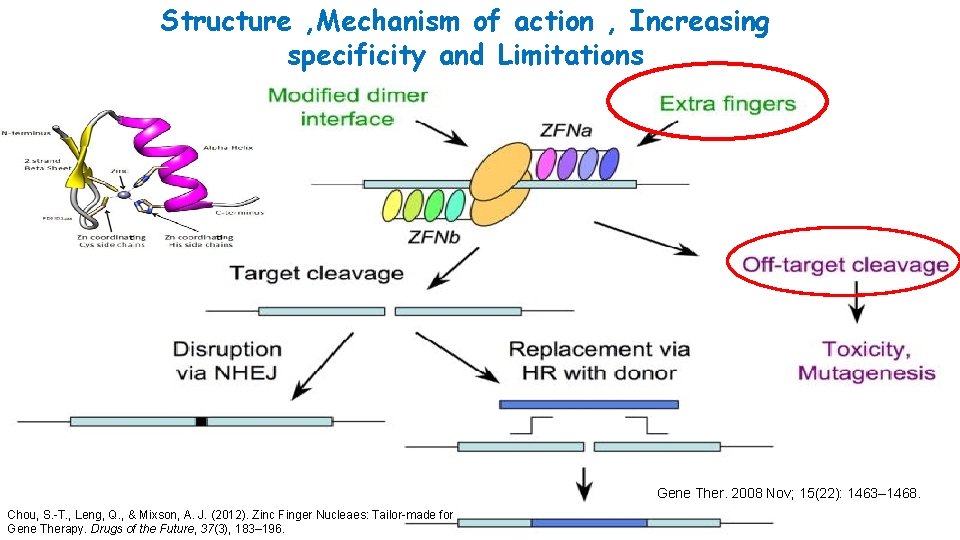
Structure , Mechanism of action , Increasing specificity and Limitations Gene Ther. 2008 Nov; 15(22): 1463– 1468. Chou, S. -T. , Leng, Q. , & Mixson, A. J. (2012). Zinc Finger Nucleaes: Tailor-made for Gene Therapy. Drugs of the Future, 37(3), 183– 196.
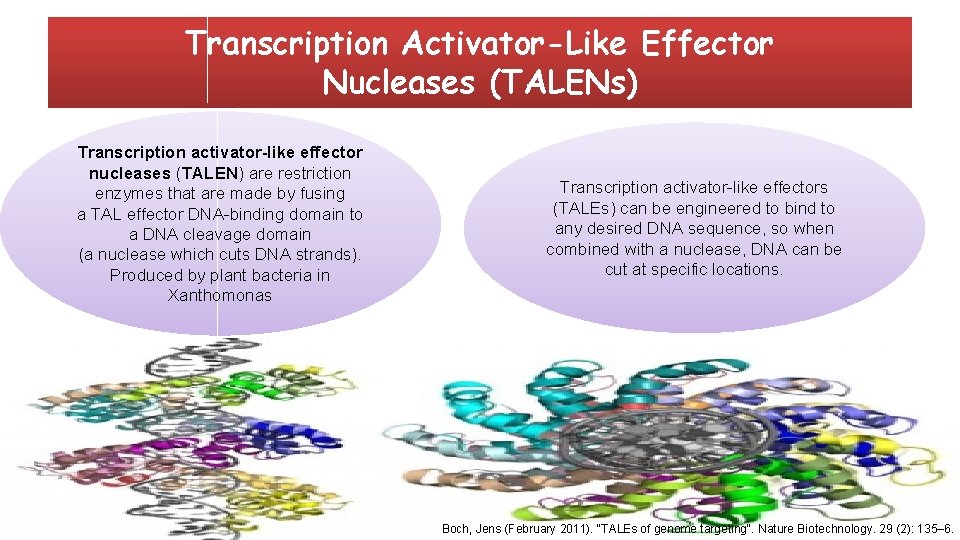
Transcription Activator-Like Effector Nucleases (TALENs) Transcription activator-like effector nucleases (TALEN) are restriction enzymes that are made by fusing a TAL effector DNA-binding domain to a DNA cleavage domain (a nuclease which cuts DNA strands). Produced by plant bacteria in Xanthomonas Transcription activator-like effectors (TALEs) can be engineered to bind to any desired DNA sequence, so when combined with a nuclease, DNA can be cut at specific locations. Boch, Jens (February 2011). "TALEs of genome targeting". Nature Biotechnology. 29 (2): 135– 6.
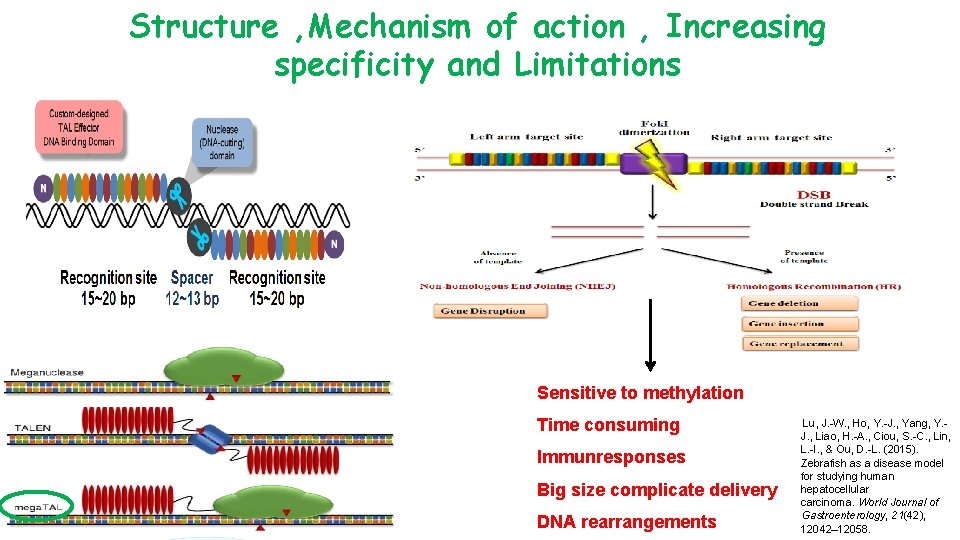
Structure , Mechanism of action , Increasing specificity and Limitations Sensitive to methylation Time consuming Immunresponses Big size complicate delivery DNA rearrangements Lu, J. -W. , Ho, Y. -J. , Yang, Y. J. , Liao, H. -A. , Ciou, S. -C. , Lin, L. -I. , & Ou, D. -L. (2015). Zebrafish as a disease model for studying human hepatocellular carcinoma. World Journal of Gastroenterology, 21(42), 12042– 12058.

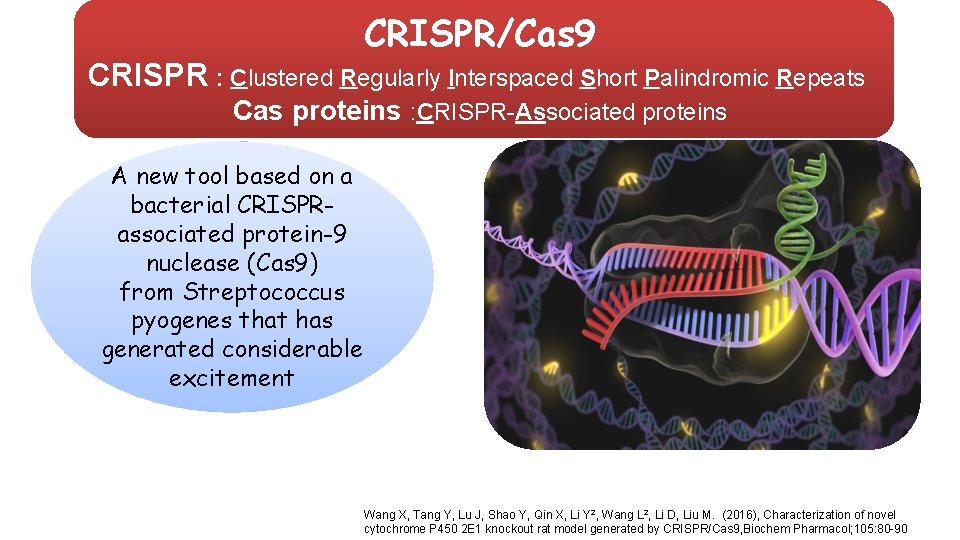
CRISPR/Cas 9 CRISPR : Clustered Regularly Interspaced Short Palindromic Repeats Cas proteins : CRISPR-Associated proteins A new tool based on a bacterial CRISPRassociated protein-9 nuclease (Cas 9) from Streptococcus pyogenes that has generated considerable excitement Wang X, Tang Y, Lu J, Shao Y, Qin X, Li Y 2, Wang L 2, Li D, Liu M. (2016), Characterization of novel cytochrome P 450 2 E 1 knockout rat model generated by CRISPR/Cas 9, Biochem Pharmacol; 105: 80 -90
What is CRISPR-Cas 9 ? CRISPR-Cas 9 is a unique technology that enables geneticists and medical researchers to edit parts of the genome by removing, adding or altering sections of the DNA sequence. It is currently the simplest, most versatile and precise method of genetic manipulation. Fangyuan Wang, Lei S. Qi (2016) Applications of CRISPR Genome Engineering in Cell Biology Volume 26, Issue 11, p 875– 888.
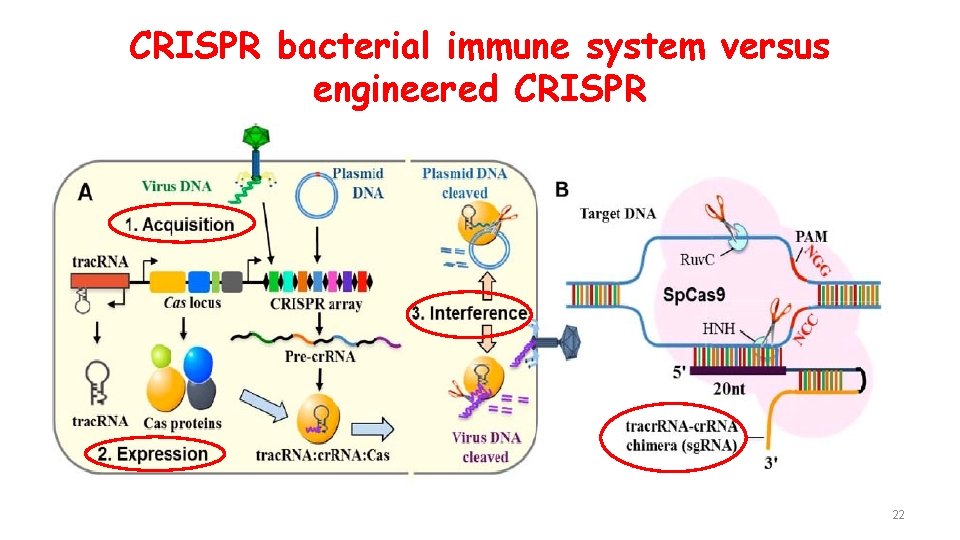
CRISPR bacterial immune system versus engineered CRISPR 22
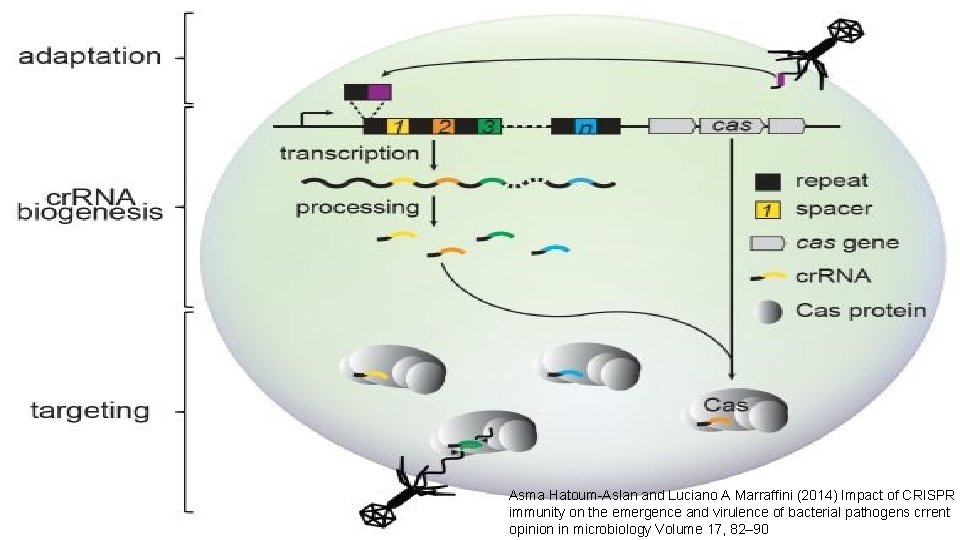
Asma Hatoum-Aslan and Luciano A Marraffini (2014) Impact of CRISPR immunity on the emergence and virulence of bacterial pathogens crrent opinion in microbiology Volume 17, 82– 90
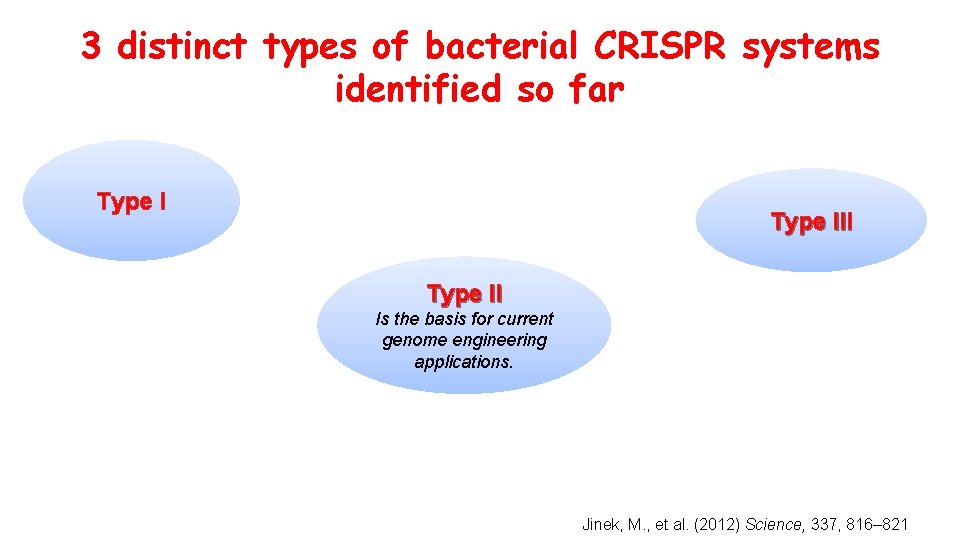
3 distinct types of bacterial CRISPR systems identified so far Type III Type II Is the basis for current genome engineering applications. Jinek, M. , et al. (2012) Science, 337, 816– 821
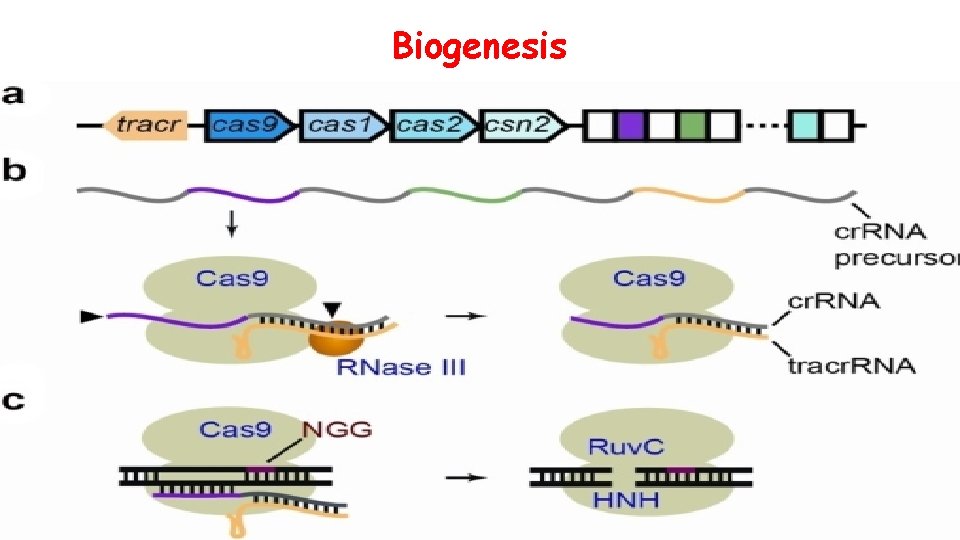
Biogenesis
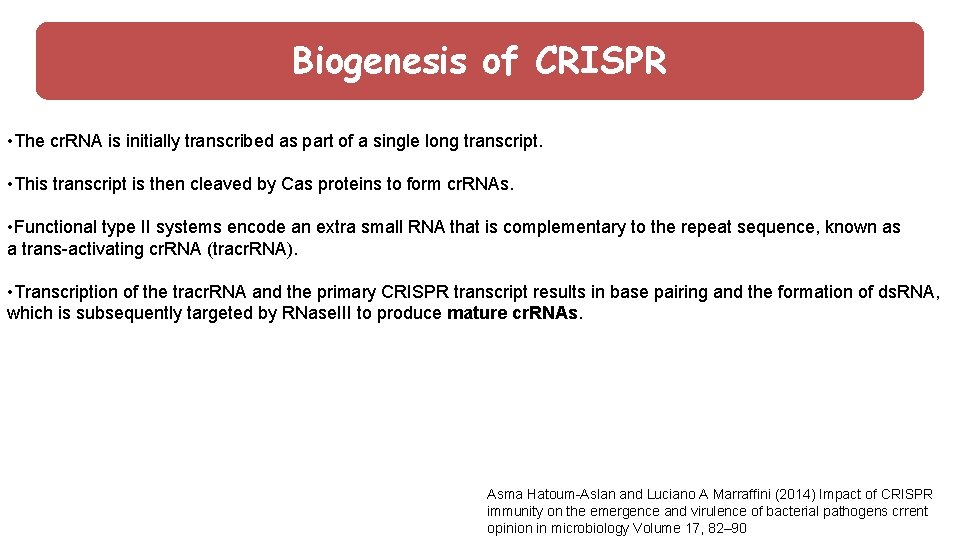
Biogenesis ofof CRISPR Biogenesis CRISPR : • The cr. RNA is initially transcribed as part of a single long transcript. • This transcript is then cleaved by Cas proteins to form cr. RNAs. • Functional type II systems encode an extra small RNA that is complementary to the repeat sequence, known as a trans-activating cr. RNA (tracr. RNA). • Transcription of the tracr. RNA and the primary CRISPR transcript results in base pairing and the formation of ds. RNA, which is subsequently targeted by RNase. III to produce mature cr. RNAs. Asma Hatoum-Aslan and Luciano A Marraffini (2014) Impact of CRISPR immunity on the emergence and virulence of bacterial pathogens crrent opinion in microbiology Volume 17, 82– 90
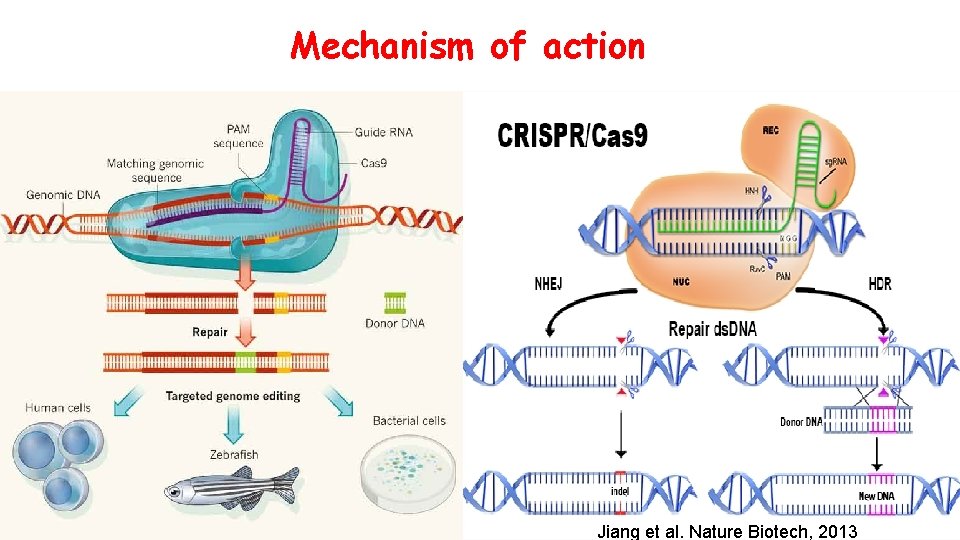
Mechanism of action Jiang et al. Nature Biotech, 2013
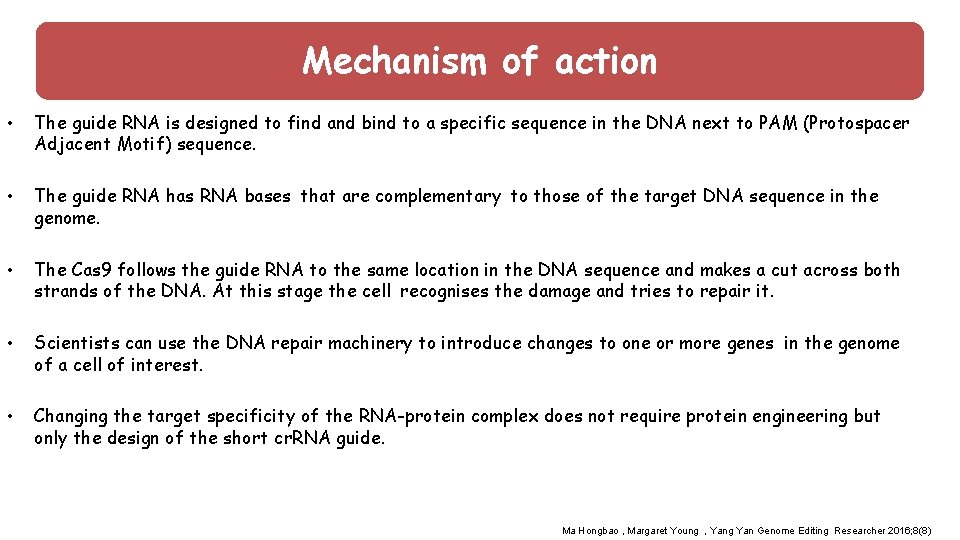
Mechanism of action : of action • The guide RNA is designed to find and bind to a specific sequence in the DNA next to PAM (Protospacer Adjacent Motif) sequence. • The guide RNA has RNA bases that are complementary to those of the target DNA sequence in the genome. • The Cas 9 follows the guide RNA to the same location in the DNA sequence and makes a cut across both strands of the DNA. At this stage the cell recognises the damage and tries to repair it. • Scientists can use the DNA repair machinery to introduce changes to one or more genes in the genome of a cell of interest. • Changing the target specificity of the RNA-protein complex does not require protein engineering but only the design of the short cr. RNA guide. Ma Hongbao , Margaret Young , Yang Yan Genome Editing Researcher 2016; 8(8)
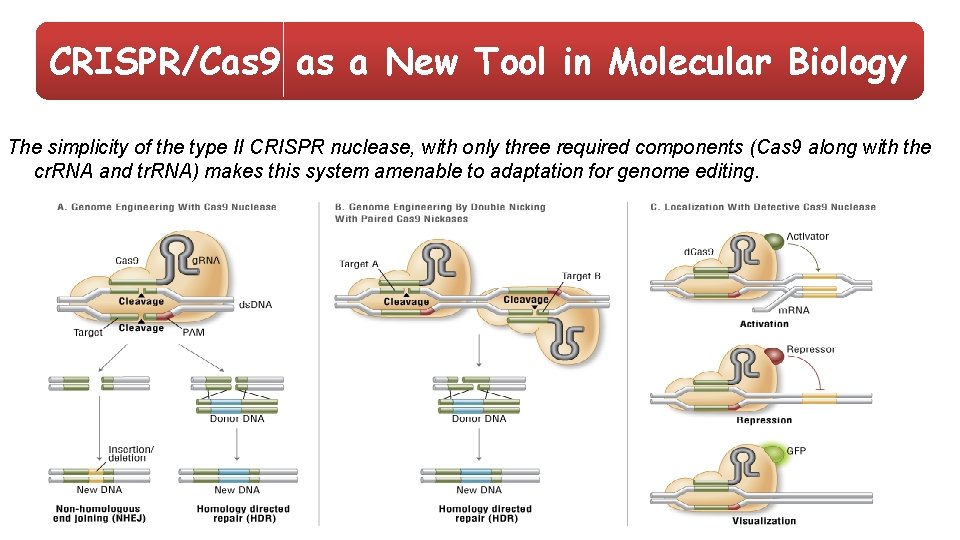
CRISPR/Cas 9 as a New Tool in Molecular Biology The simplicity of the type II CRISPR nuclease, with only three required components (Cas 9 along with the cr. RNA and tr. RNA) makes this system amenable to adaptation for genome editing.
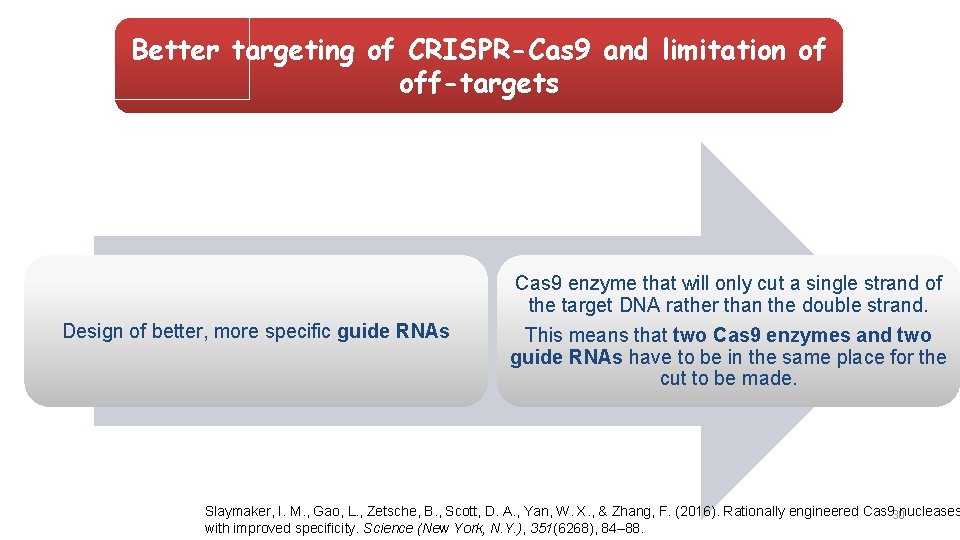
Better targeting of CRISPR-Cas 9 and limitation of off-targets Design of better, more specific guide RNAs Cas 9 enzyme that will only cut a single strand of the target DNA rather than the double strand. This means that two Cas 9 enzymes and two guide RNAs have to be in the same place for the cut to be made. Slaymaker, I. M. , Gao, L. , Zetsche, B. , Scott, D. A. , Yan, W. X. , & Zhang, F. (2016). Rationally engineered Cas 930 nucleases with improved specificity. Science (New York, N. Y. ), 351(6268), 84– 88.
• The CRISPR-Cas system was selected by Science as 2015 Breakthrough of the Year. • The CRISPR-Cas 9 system currently stands out as the fastest, cheapest and most reliable system for ‘editing’ genes.
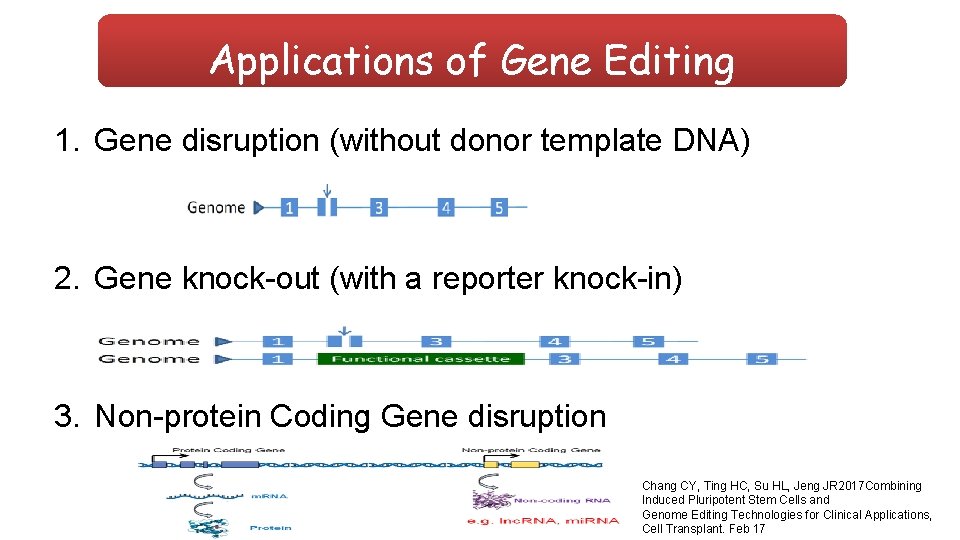
Applications of Gene Editing 1. Gene disruption (without donor template DNA) 2. Gene knock-out (with a reporter knock-in) 3. Non-protein Coding Gene disruption Chang CY, Ting HC, Su HL, Jeng JR 2017 Combining Induced Pluripotent Stem Cells and Genome Editing Technologies for Clinical Applications, Cell Transplant. Feb 17
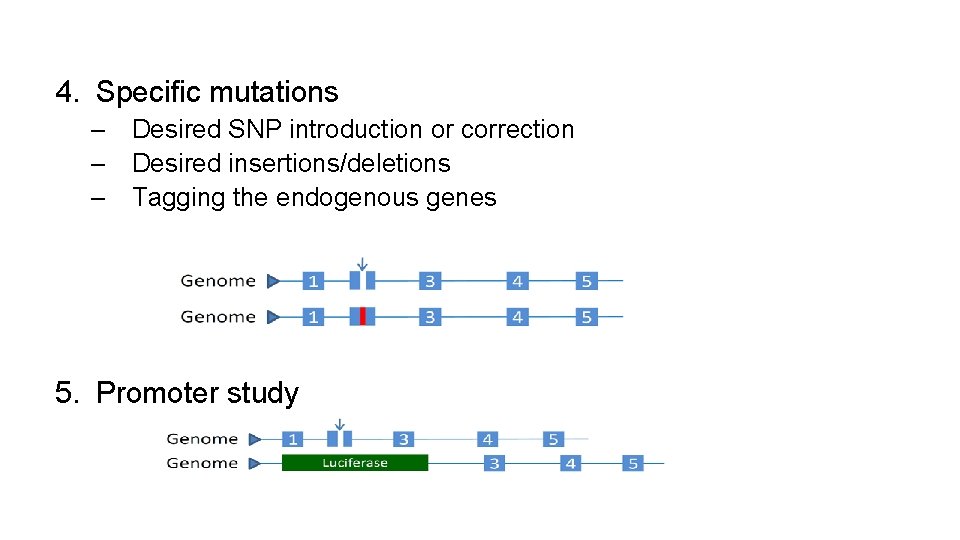
4. Specific mutations – Desired SNP introduction or correction – Desired insertions/deletions – Tagging the endogenous genes 5. Promoter study
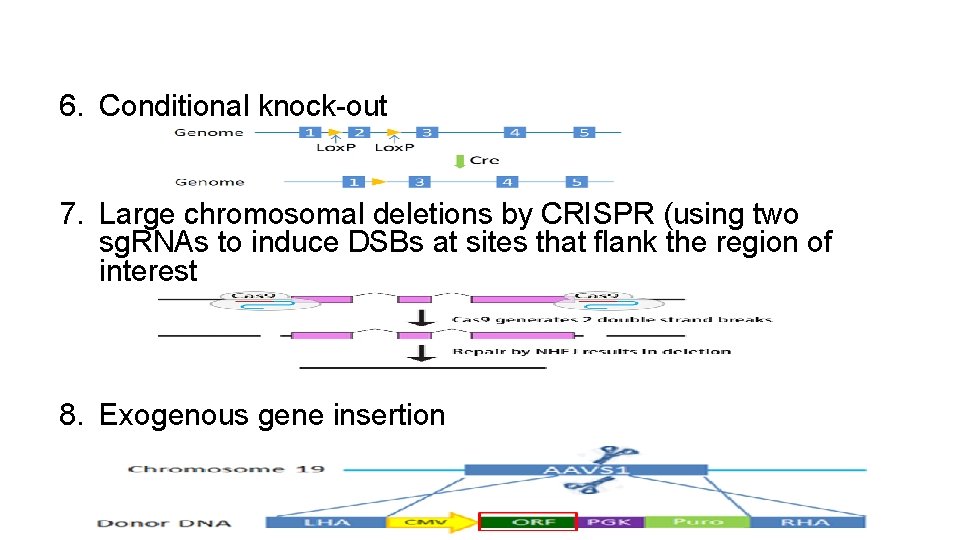
6. Conditional knock-out 7. Large chromosomal deletions by CRISPR (using two sg. RNAs to induce DSBs at sites that flank the region of interest 8. Exogenous gene insertion

Gene Editing with engineered nuclease applied in a diverse set of species

What is Gene Editing used for? For research: Gene editing can be used to change the DNA in cells or organisms to understand their biology and how they work. To treat disease: Gene editing has been used to modify human blood cells that are then put back into the body to treat conditions including leukaemia and AIDS. It could also potentially be used to treat other infections (such as MRSA) and simple genetic conditions (such as muscular dystrophy and haemophilia). Bahal, R. , Ali Mc. Neer, N. , Quijano, E. , Liu, Y. , Sulkowski, P. , Turchick, A. , … Glazer, P. M. (2016). In vivo correction of anaemia in β -thalassemic mice by γPNA-mediated gene For biotechnology: • Gene editing has been used in agriculture to genetically modify crops to improve their yields and resistance to disease and drought, as well as to genetically modify cattle that don’t have horns. Altpeter, F. , Springer, N. M. , Bartley, L. E. , Blechl, A. E. , Brutnell, T. P. , Citovsky, V. , … Stewart, C. N. (2016). Advancing Crop Transformation in the Era of Genome Editing. The Plant Cell, 28(7), 1510– 1520.

Gene Editing and Disease
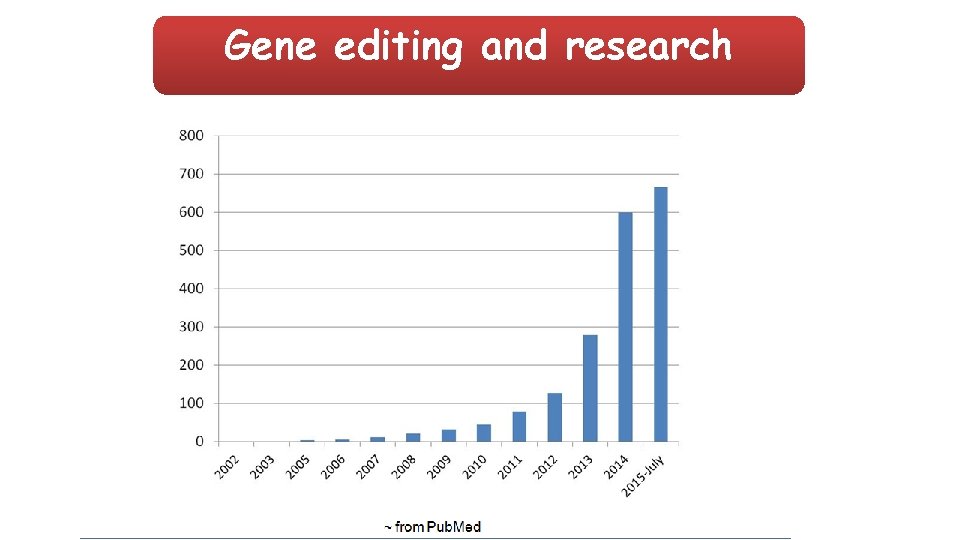
Gene editing and research


References: 1. 2. 3. Yuriko Osakabe, Keishi Osakabe; Genome Editing with Engineered Nucleases in Plants. Plant Cell Physiol 2015; 56 (3): 389 -400. Carroll, D. (2008). Zinc-finger Nucleases as Gene Therapy Agents. Gene Therapy, 15(22), 1463– 1468. Pattanayak, V. , Guilinger, J. P. , & Liu, D. R. (2014). Determining the specificities of TALENs, Cas 9, and other genome editing enzymes. Methods in Enzymology, 546, 47– 78. 4. 5. Maeder, M. L. , & Gersbach, C. A. (2016). Genome-editing Technologies for Gene and Cell Therapy. Molecular Therapy, 24(3), 430– 446. S. Arnould, C. Delenda, S. Grizot, C. Desseaux, F. Pâques, G. H. Silva, J. Smith; The I-Cre. I meganuclease and its engineered derivatives: applications from cell modification to gene therapy. Protein Eng Des Sel 2011; 24 (1 -2): 27 -31. doi: 10. 1093/protein/gzq 083 February (2009). "Selfish DNA: homing endonucleases Silva, G. , Poirot, L. , Galetto, R. , Smith, J. , Montoya, G. , Duchateau, P. , & Pâques, F. (2011). Meganucleases and Other Tools for Targeted Genome Engineering: Perspectives and Challenges for Gene Therapy. Current Gene Therapy, 11(1), 11– 27. Gao, Huirong; Smith, Jeff; Yang, Meizhu; Jones, Spencer; Djukanovic, Vesna; Nicholson, Michael G. ; West, Ande; Bidney, Dennis; Falco, S. Carl (2010). "Heritable targeted mutagenesis in maize using a designed endonuclease". The Plant Journal. 61 (1): 176– 87. 6. 7. 8. 9. 10. 11. 12. Carroll, D. (2011). Genome Engineering With Zinc-Finger Nucleases. Genetics, 188(4), 773– 782. Gene Ther. 2008 Nov; 15(22): 1463– 1468. Gaj, T. , Gersbach, C. A. , & Barbas, C. F. (2013). ZFN, TALEN and CRISPR/Cas-based methods for genome engineering. Trends in Biotechnology, 31(7), 397– 405. Chou, S. -T. , Leng, Q. , & Mixson, A. J. (2012). Zinc Finger Nucleaes: Tailor-made for Gene Therapy. Drugs of the Future, 37(3), 183– 196.

References cont… 13. 14. 25. Boch, Jens (February 2011). "TALEs of genome targeting". Nature Biotechnology. 29 (2): 135– 6. Lu, J. -W. , Ho, Y. -J. , Yang, Y. -J. , Liao, H. -A. , Ciou, S. -C. , Lin, L. -I. , & Ou, D. -L. (2015). Zebrafish as a disease model for studying human hepatocellular carcinoma. World Journal of Gastroenterology, 21(42), 12042– 12058. Joung J. K. & Sander J. D. TALENs: a widely applicable technology for targeted genome editing. Nature reviews. Mol. Cell Biol 14, 49– 55 (2013) Wang X, Tang Y, Lu J, Shao Y, Qin X, Li Y 2, Wang L 2, Li D, Liu M. (2016), Characterization of novel cytochrome P 450 2 E 1 knockout rat model generated by CRISPR/Cas 9, Biochem Pharmacol; 105: 80 -90 Fangyuan Wang, Lei S. Qi (2016) Applications of CRISPR Genome Engineering in Cell Biology Volume 26, Issue 11, p 875– 888. Asma Hatoum-Aslan and Luciano A Marraffini (2014) Impact of CRISPR immunity on the emergence and virulence of bacterial pathogens crrent opinion in microbiology Volume 17, 82– 90 Jinek, M. , et al. (2012) Science, 337, 816– 821 Slaymaker, I. M. , Gao, L. , Zetsche, B. , Scott, D. A. , Yan, W. X. , & Zhang, F. (2016). Rationally engineered Cas 9 nucleases with improved specificity. Science (New York, N. Y. ), 351(6268), 84– 88. Tycko J, Myer VE, Hsu PD. 2016 Methods for Optimizing CRISPR-Cas 9 Genome Editing Specificity, Mol Cell. ; 63(3): 355 -70. Chang CY, Ting HC, Su HL, Jeng JR 2017 Combining Induced Pluripotent Stem Cells and Genome Editing Technologies for Clinical Applications, Cell Transplant. Feb 17 Bahal, R. , Ali Mc. Neer, N. , Quijano, E. , Liu, Y. , Sulkowski, P. , Turchick, A. , … Glazer, P. M. (2016). In vivo correction of anaemia in β-thalassemic mice by γPNA-mediated gene editing with nanoparticle delivery. Nature Communications, 7, 13304. Altpeter, F. , Springer, N. M. , Bartley, L. E. , Blechl, A. E. , Brutnell, T. P. , Citovsky, V. , … Stewart, C. N. (2016). Advancing Crop Transformation in the Era of Genome Editing. The Plant Cell, 28(7), 1510– 1520. Makita Y, Hozumi H, Hotta A. Advances in genome editing technologies for treating muscular dystrophy. Clin Calcium, 2017; 27(3): 391 -399. 26. Ma Hongbao , Margaret Young , Yang Yan Genome Editing Researcher 2016; 8(8) 15. 16. 17. 18. 19. 20. 21. 22. 23. 24.

- Slides: 44