Gene 2022118 2 n n 1866 Gregor Mendel








- Slides: 47


Gene 2022/1/18 2

n n 1866 Gregor Mendel published the results of his investigations of the inheritance of "factors" in pea plants.


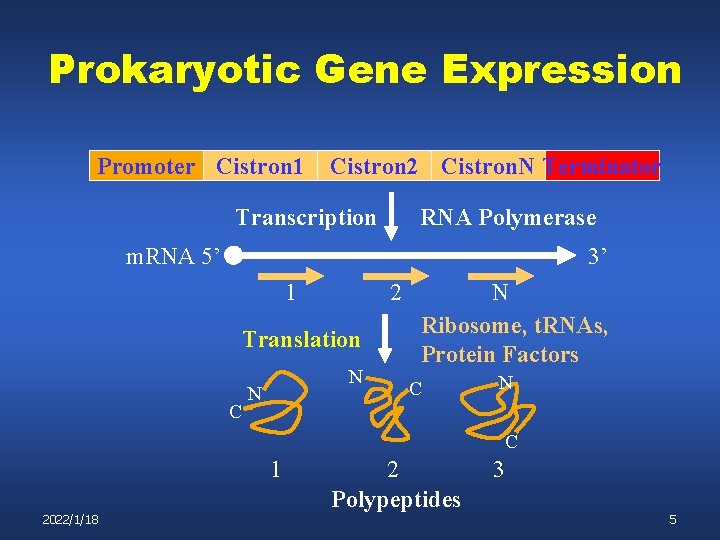
Prokaryotic Gene Expression Promoter Cistron 1 Cistron 2 Cistron. N Terminator Transcription RNA Polymerase m. RNA 5’ 3’ 1 2 Translation C N N N Ribosome, t. RNAs, Protein Factors C N C 1 2022/1/18 2 Polypeptides 3 5

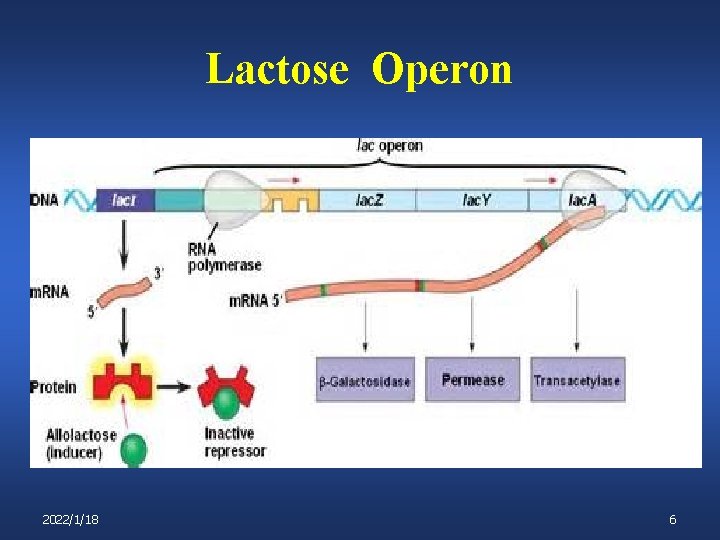
Lactose Operon 2022/1/18 6

n Pre-Transcription n Post-Transcription n Translation n Post-Translation 2022/1/18 7

Gene regulation at genome level DNA availability n Histone acetylation n DNA methylation n Genome imprinting n X chromosome inactivation 2022/1/18 8

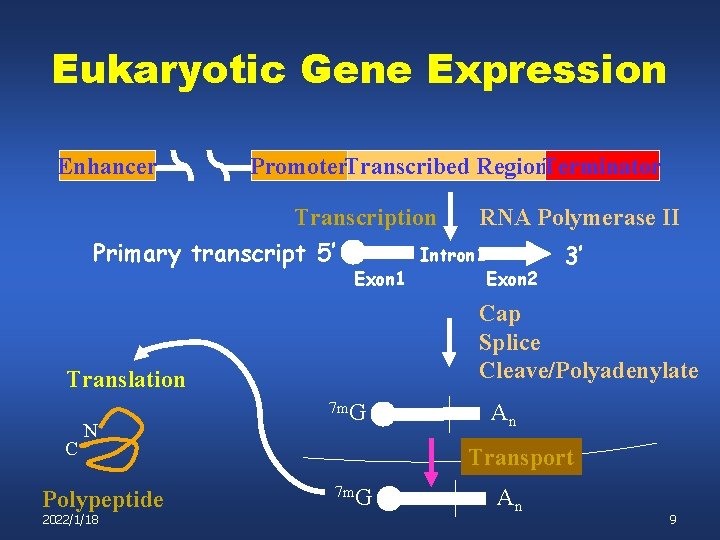
Eukaryotic Gene Expression Enhancer Promoter. Transcribed Region. Terminator Transcription RNA Polymerase II Primary transcript 5’ Intron 1 3’ Exon 1 Cap Splice Cleave/Polyadenylate Translation C N Polypeptide 2022/1/18 Exon 2 7 m. G An Transport 7 m. G An 9

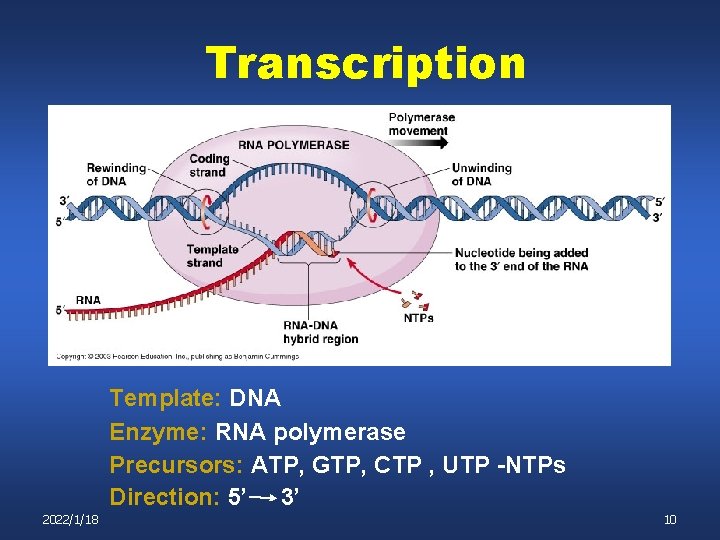
Transcription Template: DNA Enzyme: RNA polymerase Precursors: ATP, GTP, CTP , UTP -NTPs Direction: 5’ 3’ 2022/1/18 10

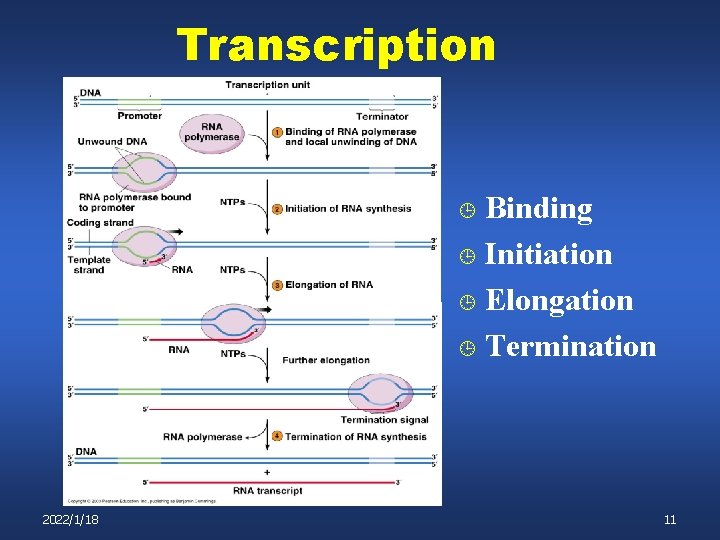
Transcription Binding ¹ Initiation ¹ Elongation ¹ Termination ¹ 2022/1/18 11

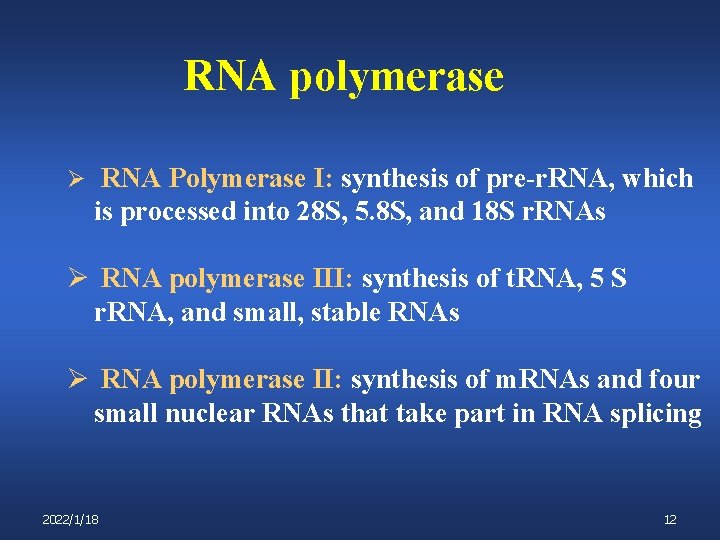
RNA polymerase Ø RNA Polymerase I: synthesis of pre-r. RNA, which is processed into 28 S, 5. 8 S, and 18 S r. RNAs Ø RNA polymerase III: synthesis of t. RNA, 5 S r. RNA, and small, stable RNAs Ø RNA polymerase II: synthesis of m. RNAs and four small nuclear RNAs that take part in RNA splicing 2022/1/18 12

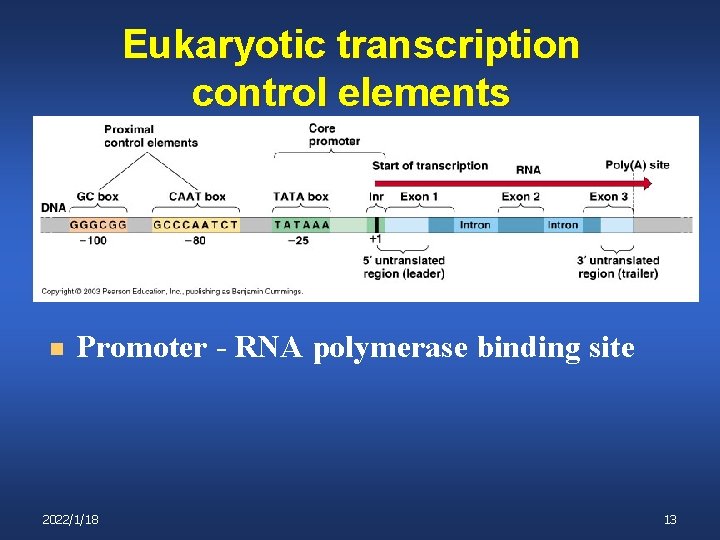
Eukaryotic transcription control elements n Promoter - RNA polymerase binding site 2022/1/18 13

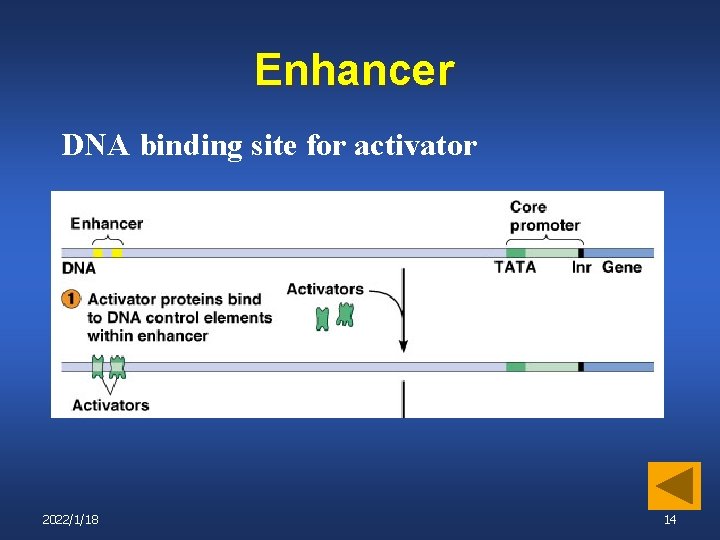
Enhancer DNA binding site for activator 2022/1/18 14

Transcription factors v v Any protein required for initiation but not a component of RNA polymerase is defined as a transcription factor. Many bind cis-acting elements in upstream region or enhancer. Some bind to other factors bound to upstream region or enhancer. Common mode of regulation is positive: activation of promoter. 2022/1/18 15

Transcription factors Activators stimulate transcription v Repressors inhibit transcription v Composed of two parts v § DNA-binding domain § Activation (or repression) domain 2022/1/18 16

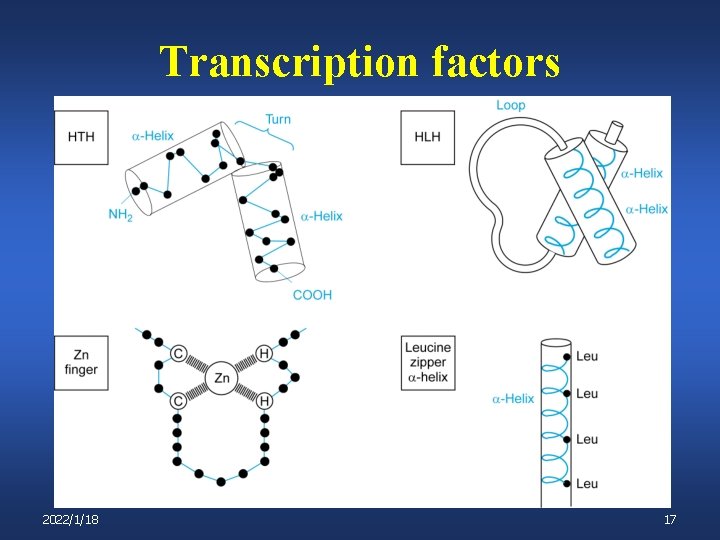
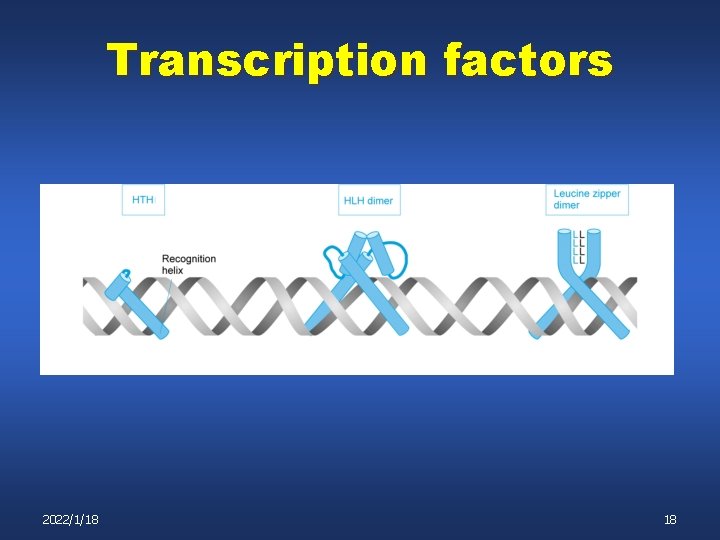
Transcription factors 2022/1/18 17

Transcription factors 2022/1/18 18

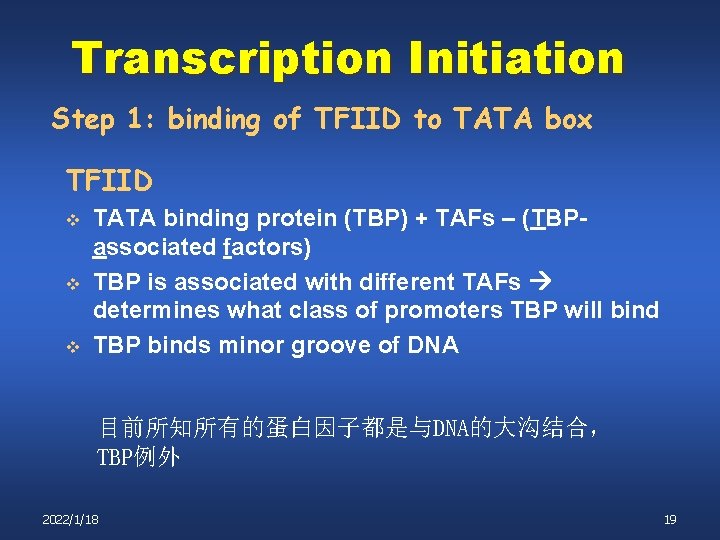
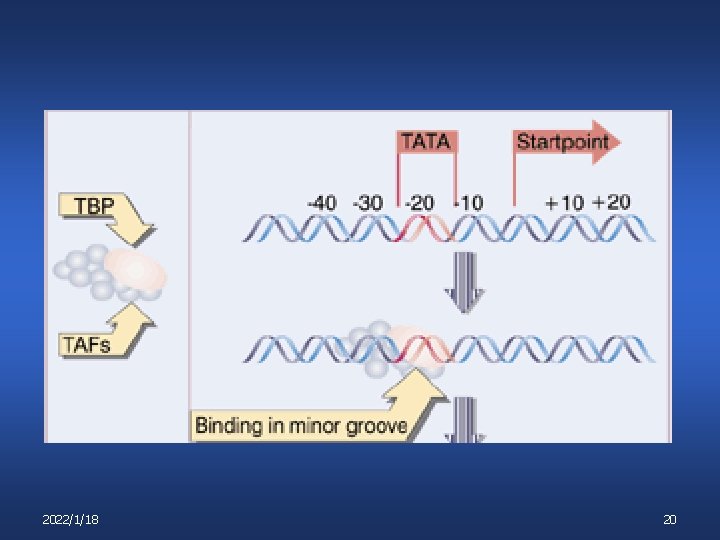
Transcription Initiation Step 1: binding of TFIID to TATA box TFIID v v v TATA binding protein (TBP) + TAFs – (TBPassociated factors) TBP is associated with different TAFs determines what class of promoters TBP will bind TBP binds minor groove of DNA 目前所知所有的蛋白因子都是与DNA的大沟结合, TBP例外 2022/1/18 19

2022/1/18 20

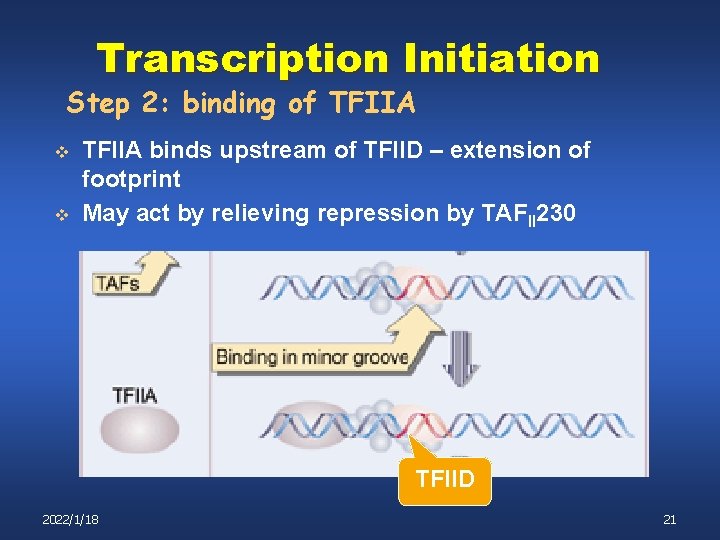
Transcription Initiation Step 2: binding of TFIIA v v TFIIA binds upstream of TFIID – extension of footprint May act by relieving repression by TAFII 230 TFIID 2022/1/18 21

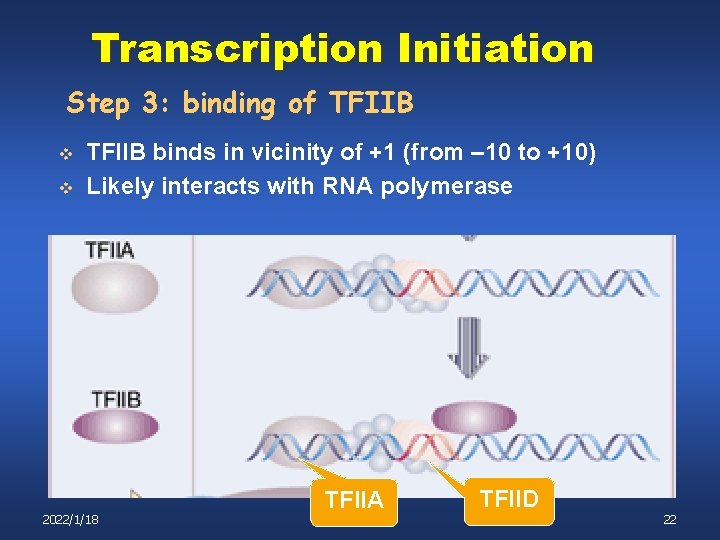
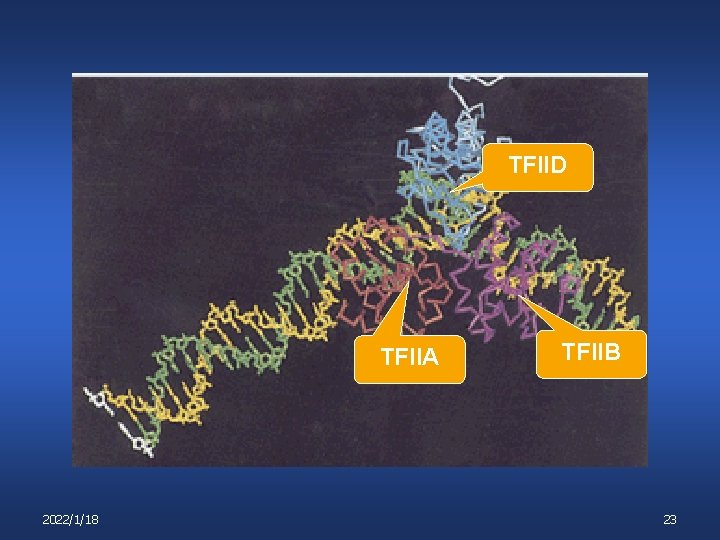
Transcription Initiation Step 3: binding of TFIIB v v TFIIB binds in vicinity of +1 (from – 10 to +10) Likely interacts with RNA polymerase 2022/1/18 TFIIA TFIID 22

TFIID TFIIA 2022/1/18 TFIIB 23

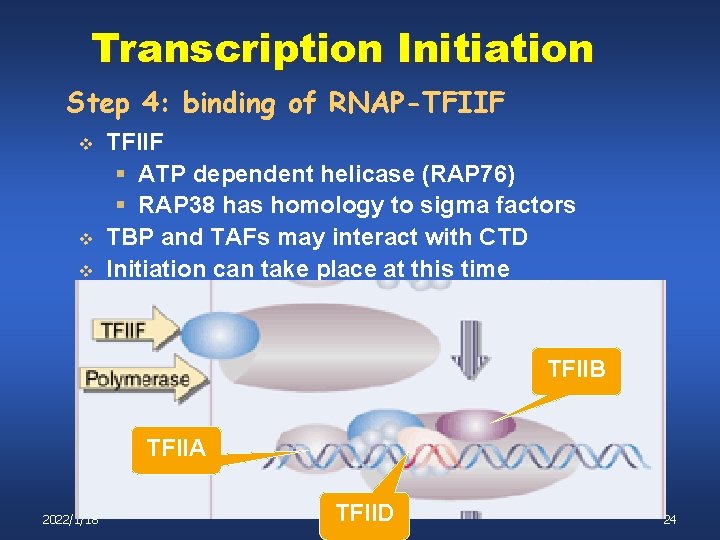
Transcription Initiation Step 4: binding of RNAP-TFIIF v v v TFIIF § ATP dependent helicase (RAP 76) § RAP 38 has homology to sigma factors TBP and TAFs may interact with CTD Initiation can take place at this time TFIIB TFIIA 2022/1/18 TFIID 24

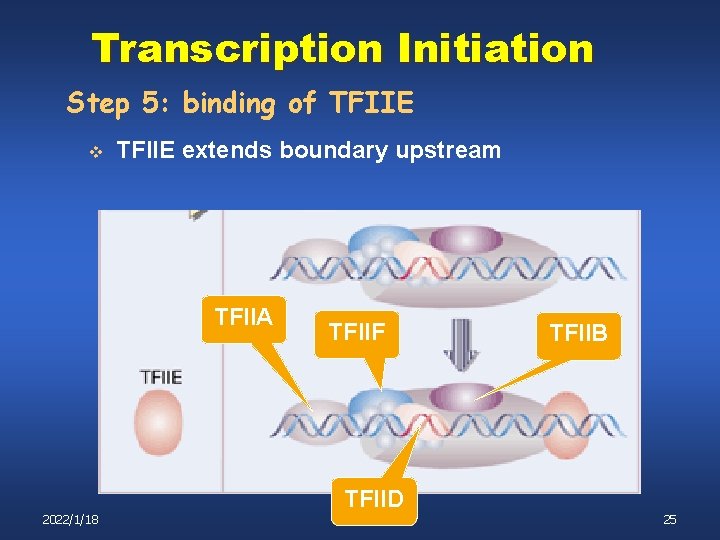
Transcription Initiation Step 5: binding of TFIIE v TFIIE extends boundary upstream TFIIA TFIIF TFIIB TFIID 2022/1/18 25

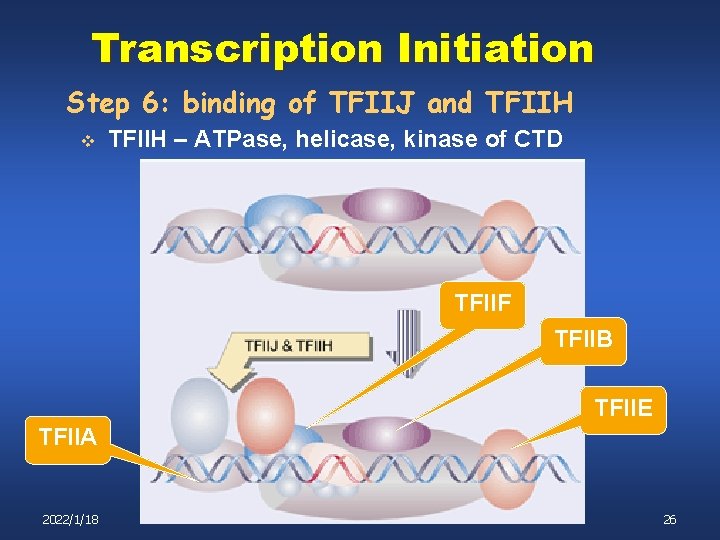
Transcription Initiation Step 6: binding of TFIIJ and TFIIH v TFIIH – ATPase, helicase, kinase of CTD TFIIF TFIIB TFIIE TFIIA 2022/1/18 26

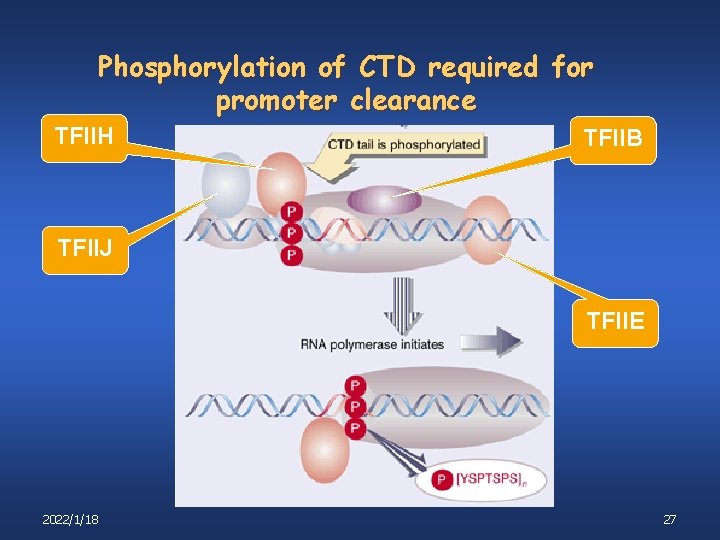
Phosphorylation of CTD required for promoter clearance TFIIH TFIIB TFIIJ TFIIE 2022/1/18 27

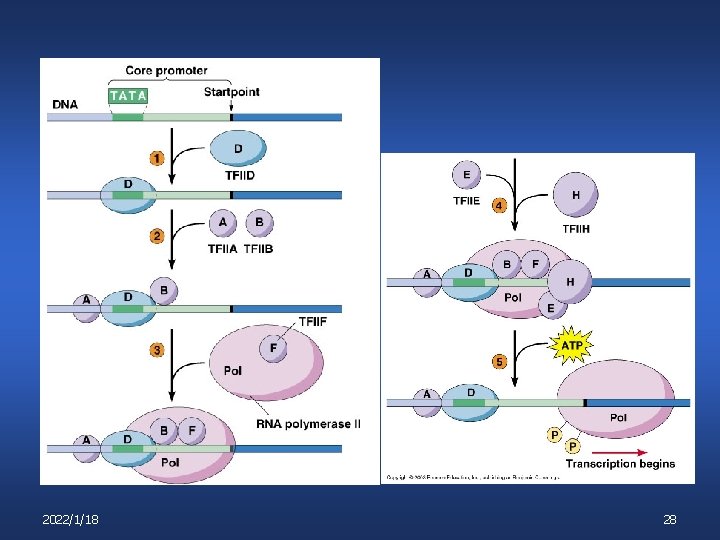
2022/1/18 28

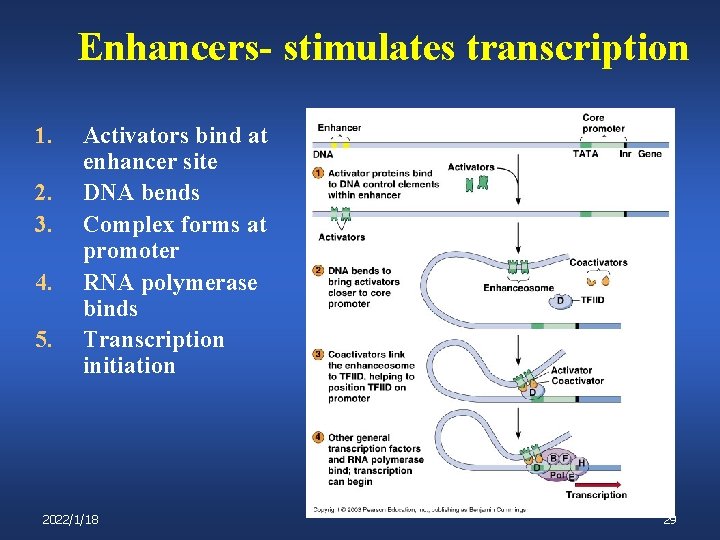
Enhancers- stimulates transcription 1. 2. 3. 4. 5. Activators bind at enhancer site DNA bends Complex forms at promoter RNA polymerase binds Transcription initiation 2022/1/18 29

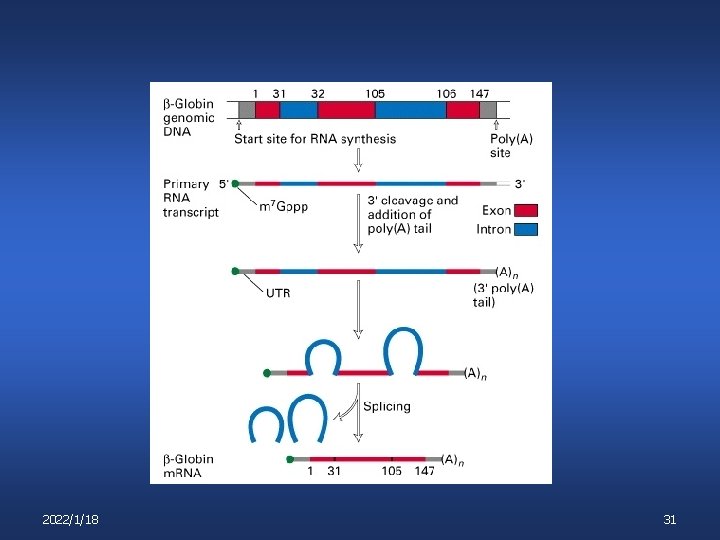
RNA processing 6 6 6 2022/1/18 Capping Polyadenylation Splicing Regulated splicing Editing 30

2022/1/18 31

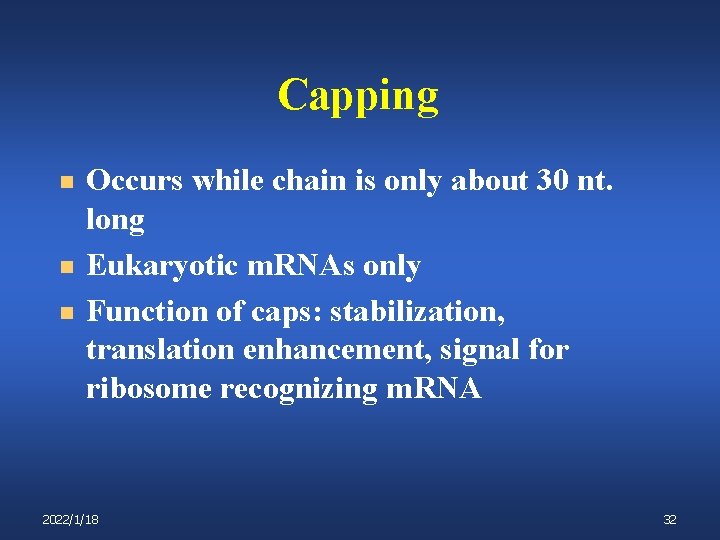
Capping n n n Occurs while chain is only about 30 nt. long Eukaryotic m. RNAs only Function of caps: stabilization, translation enhancement, signal for ribosome recognizing m. RNA 2022/1/18 32

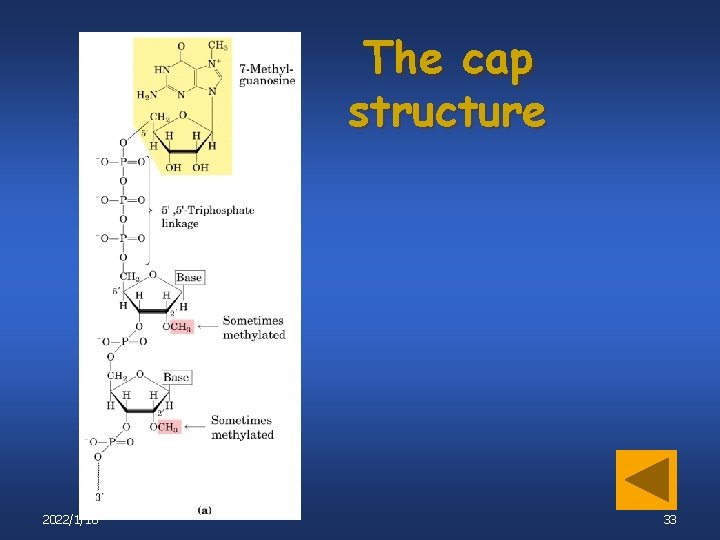
The cap structure 2022/1/18 33

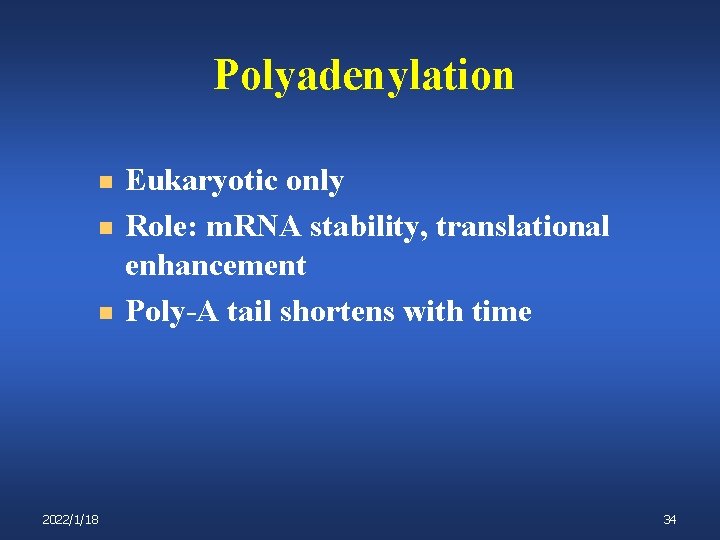
Polyadenylation n 2022/1/18 Eukaryotic only Role: m. RNA stability, translational enhancement Poly-A tail shortens with time 34

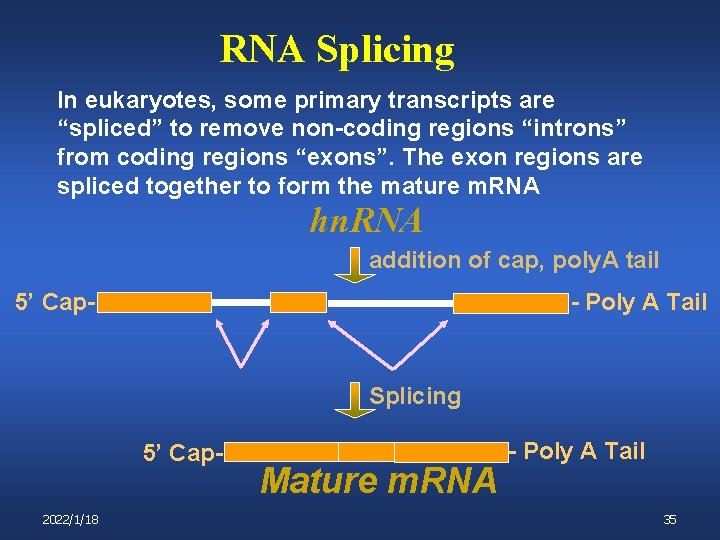
RNA Splicing In eukaryotes, some primary transcripts are “spliced” to remove non-coding regions “introns” from coding regions “exons”. The exon regions are spliced together to form the mature m. RNA hn. RNA addition of cap, poly. A tail 5’ Cap- - Poly A Tail Splicing 5’ Cap 2022/1/18 Mature m. RNA - Poly A Tail 35

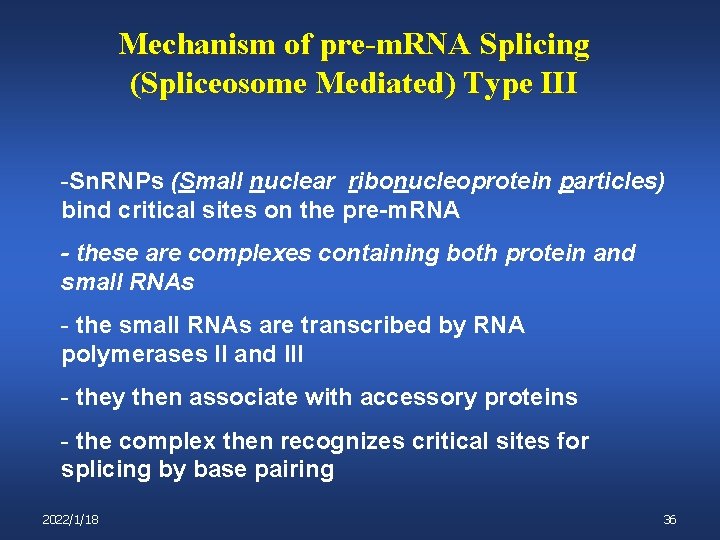
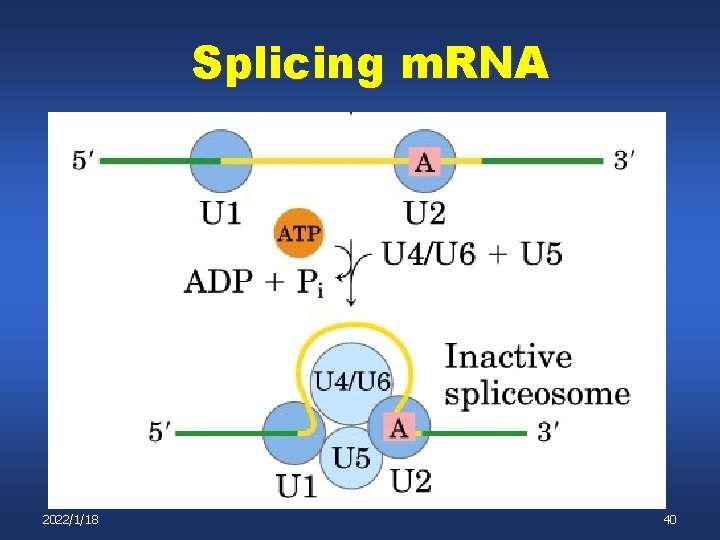
Mechanism of pre-m. RNA Splicing (Spliceosome Mediated) Type III -Sn. RNPs (Small nuclear ribonucleoprotein particles) bind critical sites on the pre-m. RNA - these are complexes containing both protein and small RNAs - the small RNAs are transcribed by RNA polymerases II and III - they then associate with accessory proteins - the complex then recognizes critical sites for splicing by base pairing 2022/1/18 36

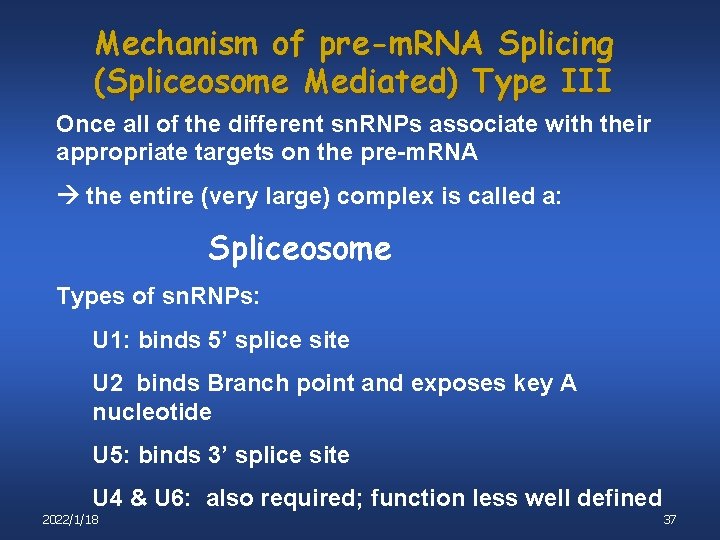
Mechanism of pre-m. RNA Splicing (Spliceosome Mediated) Type III Once all of the different sn. RNPs associate with their appropriate targets on the pre-m. RNA the entire (very large) complex is called a: Spliceosome Types of sn. RNPs: U 1: binds 5’ splice site U 2 binds Branch point and exposes key A nucleotide U 5: binds 3’ splice site U 4 & U 6: also required; function less well defined 2022/1/18 37

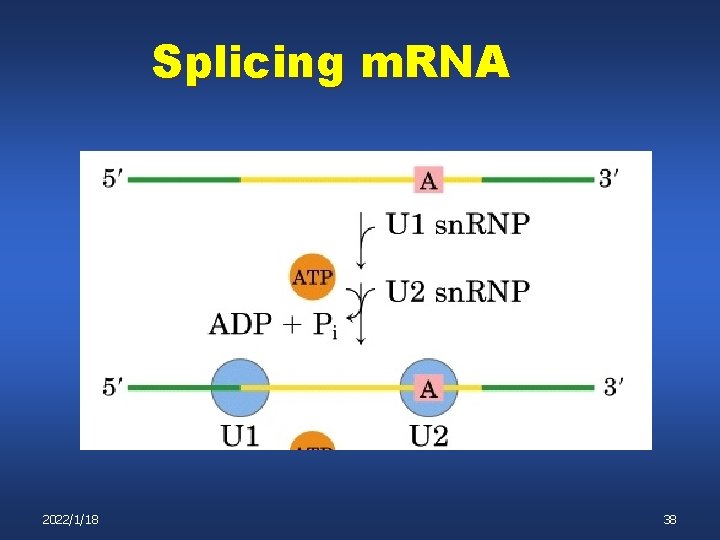
Splicing m. RNA 2022/1/18 38

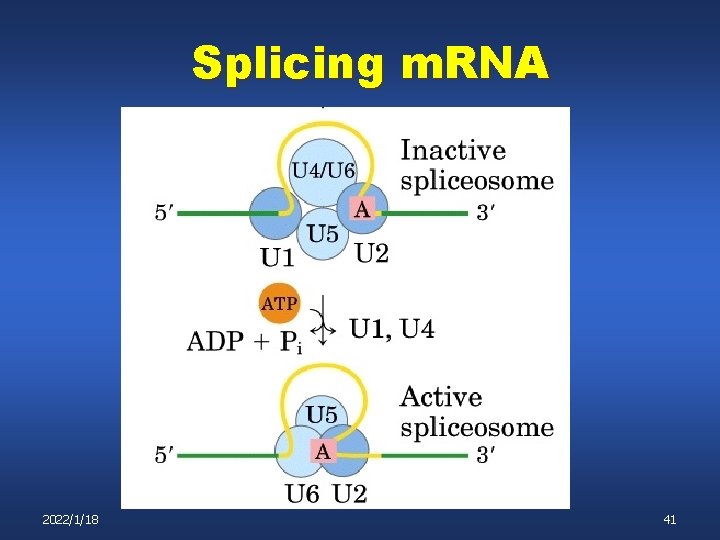
Splicing m. RNA U 4, U 5, U 6 complex binds n n U 4 and U 6 are base paired to each other After spliceosome assembly the U 4 U 6 base pairing is lost 2022/1/18 39

Splicing m. RNA 2022/1/18 40

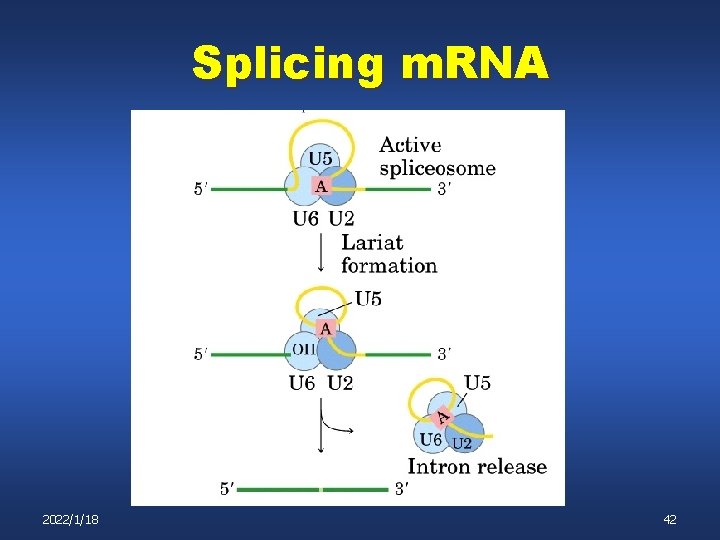
Splicing m. RNA 2022/1/18 41

Splicing m. RNA 2022/1/18 42

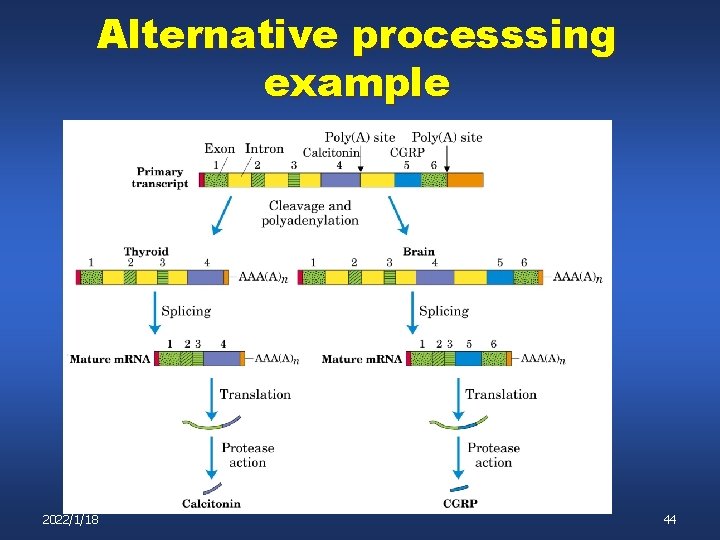
Alternative splicing is common n n Single primary transcript may be spliced into several m. RNAs. The alternatively-spliced products may be regulated 2022/1/18 43

Alternative processsing example 2022/1/18 44

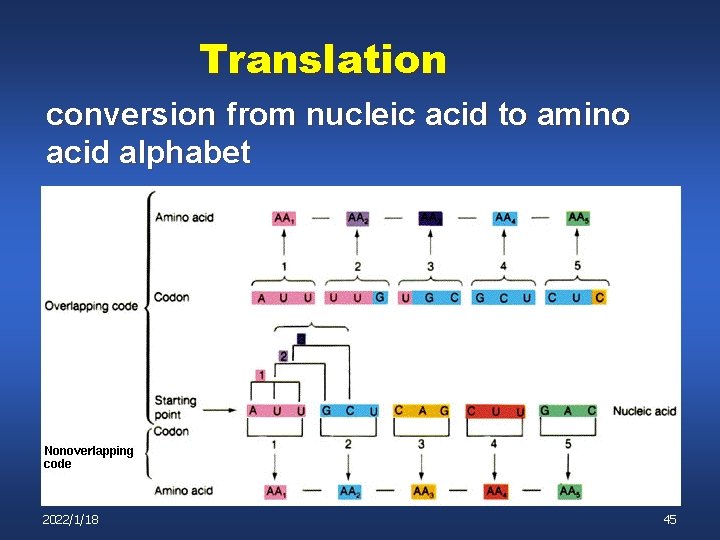
Translation conversion from nucleic acid to amino acid alphabet 2022/1/18 45

Translational control of gene expression n m. RNA noncoding segments – untranslated regions (UTRs) 5’ – UTR – from methylguanosine cap to AUG initiation codon n 3’ – UTR – from termination codon to end of poly(A) tail n n UTRs contain nucleotide sequences which mediate translational-level control 2022/1/18 46

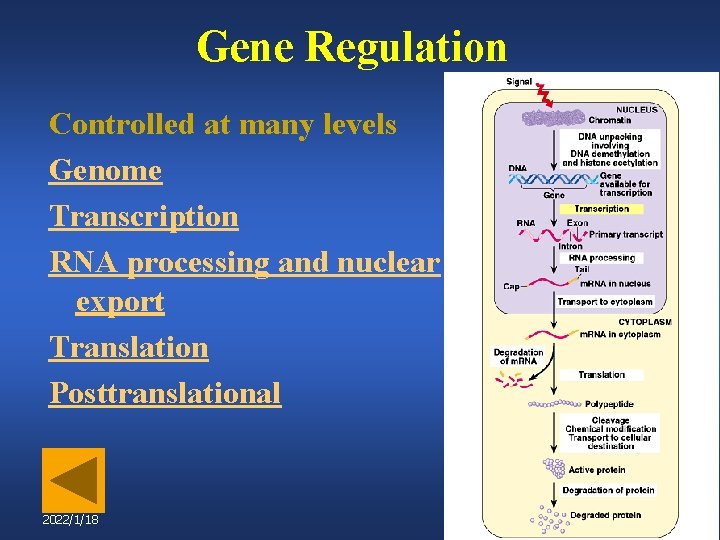
Gene Regulation Controlled at many levels Genome Transcription RNA processing and nuclear export Translation Posttranslational 2022/1/18 47