Gen ASM A High Performance LowPower Approximate String



- Slides: 52

Gen. ASM: A High Performance, Low-Power Approximate String Matching Acceleration Framework for Genome Sequence Analysis Damla Senol Cali Carnegie Mellon University (dsenol@andrew. cmu. edu) Gurpreet S. Kalsi 2, Zulal Bingol 3, Can Firtina 4, Lavanya Subramanian 5, Jeremie Kim 1, 4, Rachata Ausavarungnirun 6, 1, Mohammed Alser 4, Juan Gomez-Luna 4, Amirali Boroumand 1, Anant Nori 2, Allison Scibisz 1, Sreenivas Subramoney 2, Can Alkan 3, Saugata Ghose 7, 1, and Onur Mutlu 4, 1, 3 1 2 5 6 3 4 7 1, 4

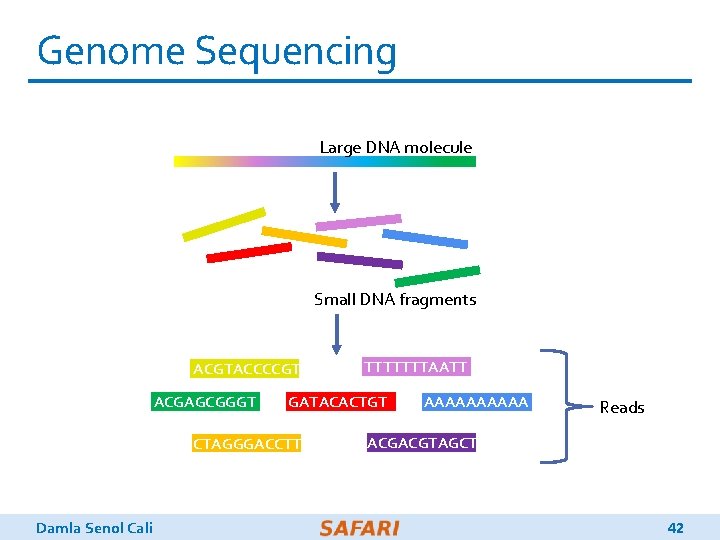
Genome Sequencing q Genome sequencing: Enables us to determine the order of the DNA sequence in an organism’s genome o Plays a pivotal role in: § Personalized medicine § Outbreak tracing § Understanding of evolution q Modern genome sequencing machines extract smaller randomized fragments of the original DNA sequence, known as reads o Short reads: a few hundred base pairs, error rate of ∼ 0. 1% o Long reads: thousands to millions of base pairs, error rate of 10– 15% Damla Senol Cali 2

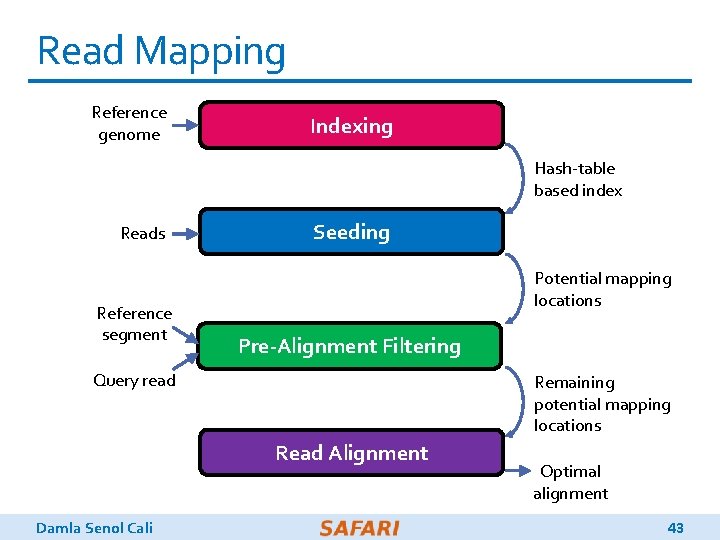
Genome Sequence Analysis q Read mapping: First key step in genome sequence analysis (GSA) o Aligns reads to one or more possible locations within the reference genome, and o Finds the matches and differences between the read and the reference genome segment at that location q Multiple steps of read mapping require approximate string matching o Approximate string matching (ASM) enables read mapping to account for sequencing errors and genetic variations in the reads q Bottlenecked by the computational power and memory bandwidth limitations of existing systems Damla Senol Cali 3

Gen. ASM: ASM Framework for GSA Our Goal: Accelerate approximate string matching by designing a fast and flexible framework, which can accelerate multiple steps of genome sequence analysis q Gen. ASM: First ASM acceleration framework for GSA o Based upon the Bitap algorithm § Uses fast and simple bitwise operations to perform ASM o Modified and extended ASM algorithm § Highly-parallel Bitap with long read support § Novel bitvector-based algorithm to perform traceback o Co-design of our modified scalable and memory-efficient algorithms with low-power and area-efficient hardware accelerators Damla Senol Cali 4

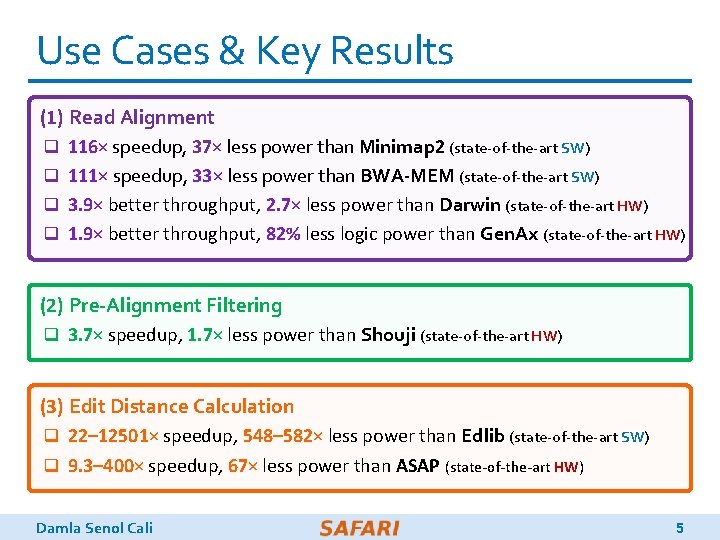
Use Cases & Key Results (1) Read Alignment q 116× speedup, 37× less power than Minimap 2 (state-of-the-art SW) q 111× speedup, 33× less power than BWA-MEM (state-of-the-art SW) q 3. 9× better throughput, 2. 7× less power than Darwin (state-of-the-art HW) q 1. 9× better throughput, 82% less logic power than Gen. Ax (state-of-the-art HW) (2) Pre-Alignment Filtering q 3. 7× speedup, 1. 7× less power than Shouji (state-of-the-art HW) (3) Edit Distance Calculation q 22– 12501× speedup, 548– 582× less power than Edlib (state-of-the-art SW) q 9. 3– 400× speedup, 67× less power than ASAP (state-of-the-art HW) Damla Senol Cali 5

Outline q Introduction q Background o Approximate String Matching (ASM) o ASM with Bitap Algorithm q Gen. ASM: ASM Acceleration Framework o Gen. ASM Algorithm o Gen. ASM Hardware Design o Use Cases of Gen. ASM q Evaluation q Conclusion Damla Senol Cali 6

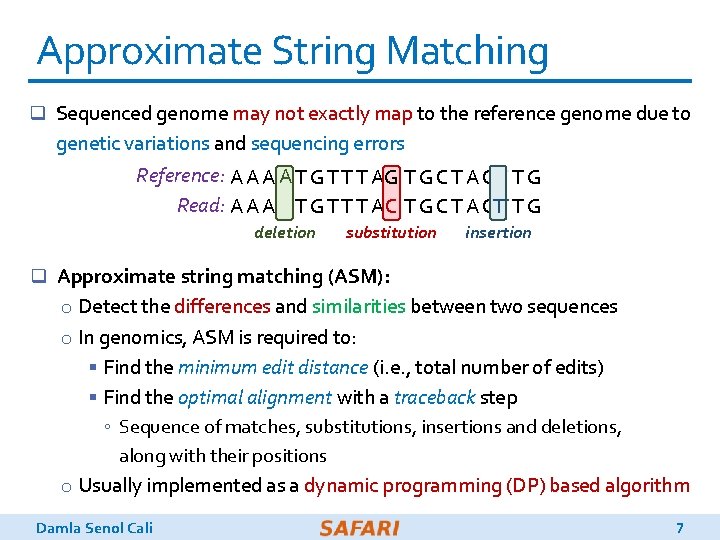
Approximate String Matching q Sequenced genome may not exactly map to the reference genome due to genetic variations and sequencing errors Reference: A A T G T T T AG GTGCTACTG TG Read: A A A T CG TG TC A TCTT G TG TTTA T C TC A AG TG TC AC G deletion substitution insertion q Approximate string matching (ASM): o Detect the differences and similarities between two sequences o In genomics, ASM is required to: § Find the minimum edit distance (i. e. , total number of edits) § Find the optimal alignment with a traceback step ◦ Sequence of matches, substitutions, insertions and deletions, along with their positions o Usually implemented as a dynamic programming (DP) based algorithm Damla Senol Cali 7

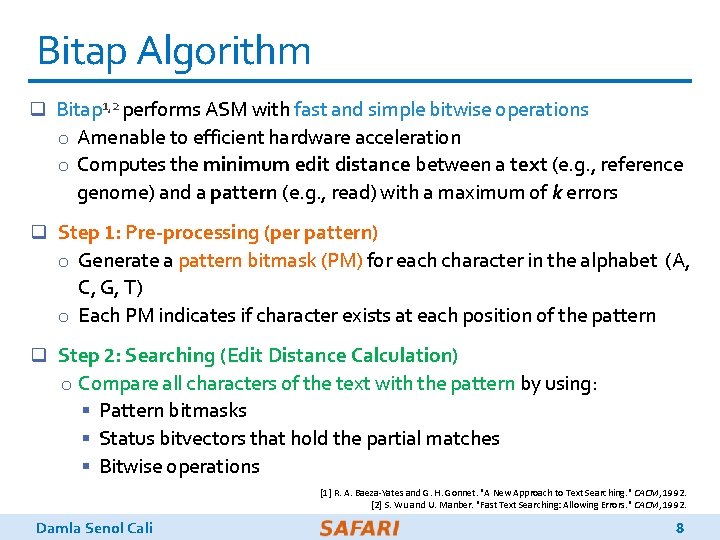
Bitap Algorithm q Bitap 1, 2 performs ASM with fast and simple bitwise operations o Amenable to efficient hardware acceleration o Computes the minimum edit distance between a text (e. g. , reference genome) and a pattern (e. g. , read) with a maximum of k errors q Step 1: Pre-processing (per pattern) o Generate a pattern bitmask (PM) for each character in the alphabet (A, C, G, T) o Each PM indicates if character exists at each position of the pattern q Step 2: Searching (Edit Distance Calculation) o Compare all characters of the text with the pattern by using: § Pattern bitmasks § Status bitvectors that hold the partial matches § Bitwise operations [1] R. A. Baeza-Yates and G. H. Gonnet. "A New Approach to Text Searching. " CACM, 1992. [2] S. Wu and U. Manber. "Fast Text Searching: Allowing Errors. " CACM, 1992. Damla Senol Cali 8

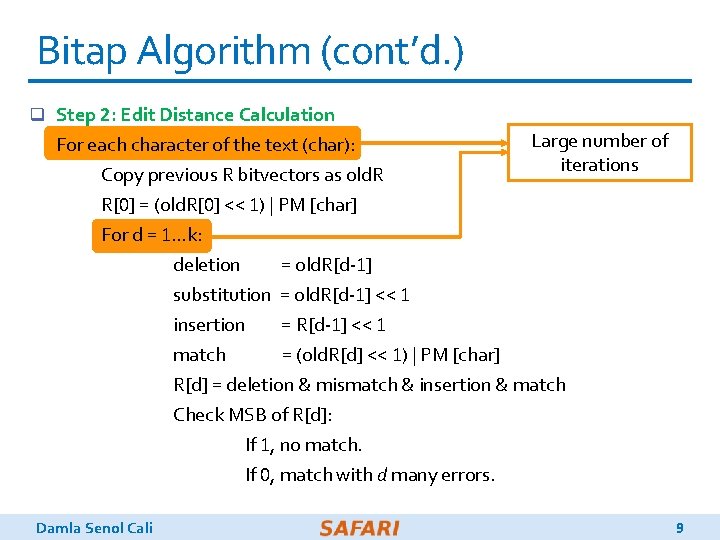
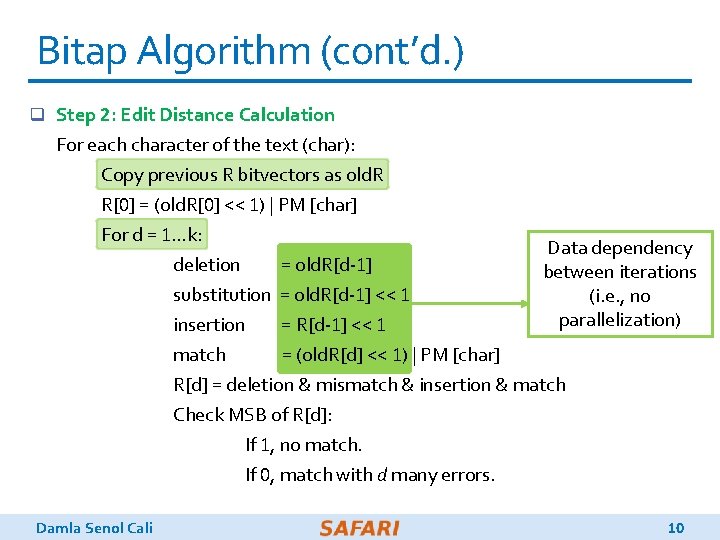
Bitap Algorithm (cont’d. ) q Step 2: Edit Distance Calculation Large number of For each character of the text (char): iterations Copy previous R bitvectors as old. R R[0] = (old. R[0] << 1) | PM [char] For d = 1…k: deletion = old. R[d-1] substitution = old. R[d-1] << 1 insertion = R[d-1] << 1 match = (old. R[d] << 1) | PM [char] R[d] = deletion & mismatch & insertion & match Check MSB of R[d]: If 1, no match. If 0, match with d many errors. Damla Senol Cali 9

Bitap Algorithm (cont’d. ) q Step 2: Edit Distance Calculation For each character of the text (char): Copy previous R bitvectors as old. R R[0] = (old. R[0] << 1) | PM [char] For d = 1…k: Data dependency deletion = old. R[d-1] between iterations substitution = old. R[d-1] << 1 (i. e. , no parallelization) insertion = R[d-1] << 1 match = (old. R[d] << 1) | PM [char] R[d] = deletion & mismatch & insertion & match Check MSB of R[d]: If 1, no match. If 0, match with d many errors. Damla Senol Cali 10

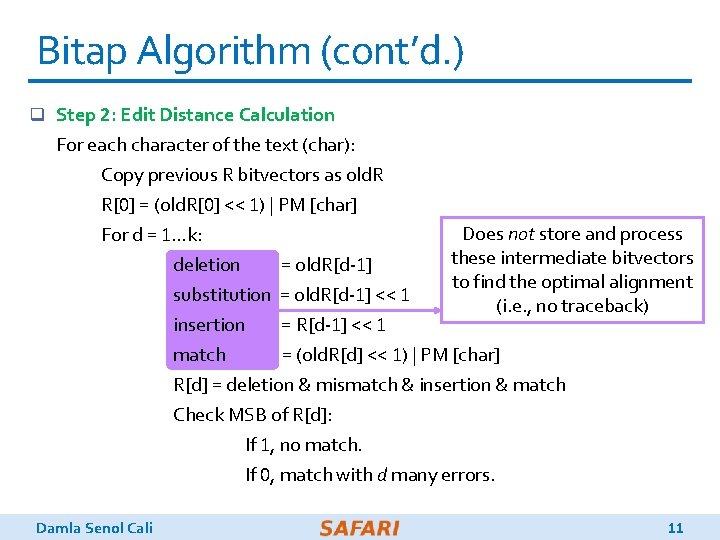
Bitap Algorithm (cont’d. ) q Step 2: Edit Distance Calculation For each character of the text (char): Copy previous R bitvectors as old. R R[0] = (old. R[0] << 1) | PM [char] Does not store and process For d = 1…k: these intermediate bitvectors deletion = old. R[d-1] to find the optimal alignment substitution = old. R[d-1] << 1 (i. e. , no traceback) insertion = R[d-1] << 1 match = (old. R[d] << 1) | PM [char] R[d] = deletion & mismatch & insertion & match Check MSB of R[d]: If 1, no match. If 0, match with d many errors. Damla Senol Cali 11

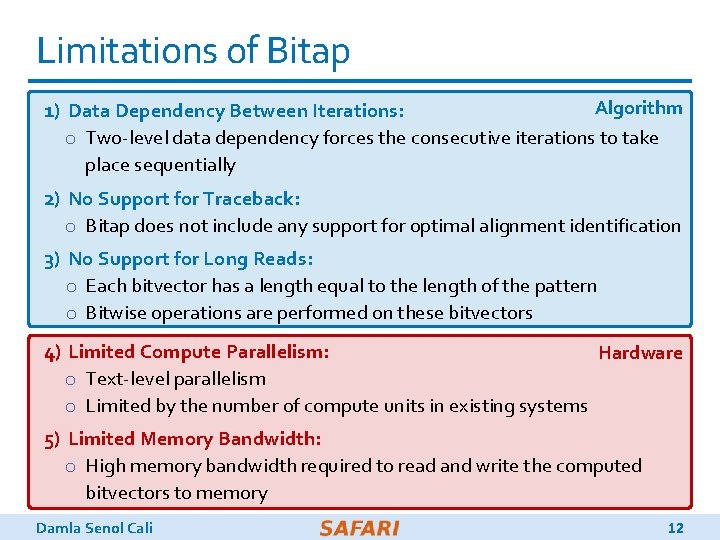
Limitations of Bitap Algorithm 1) Data Dependency Between Iterations: o Two-level data dependency forces the consecutive iterations to take place sequentially 2) No Support for Traceback: o Bitap does not include any support for optimal alignment identification 3) No Support for Long Reads: o Each bitvector has a length equal to the length of the pattern o Bitwise operations are performed on these bitvectors 4) Limited Compute Parallelism: Hardware o Text-level parallelism o Limited by the number of compute units in existing systems 5) Limited Memory Bandwidth: o High memory bandwidth required to read and write the computed bitvectors to memory Damla Senol Cali 12

Outline q Introduction q Background o Approximate String Matching (ASM) o ASM with Bitap Algorithm q Gen. ASM: ASM Acceleration Framework o Gen. ASM Algorithm o Gen. ASM Hardware Design o Use Cases of Gen. ASM q Evaluation q Conclusion Damla Senol Cali 13

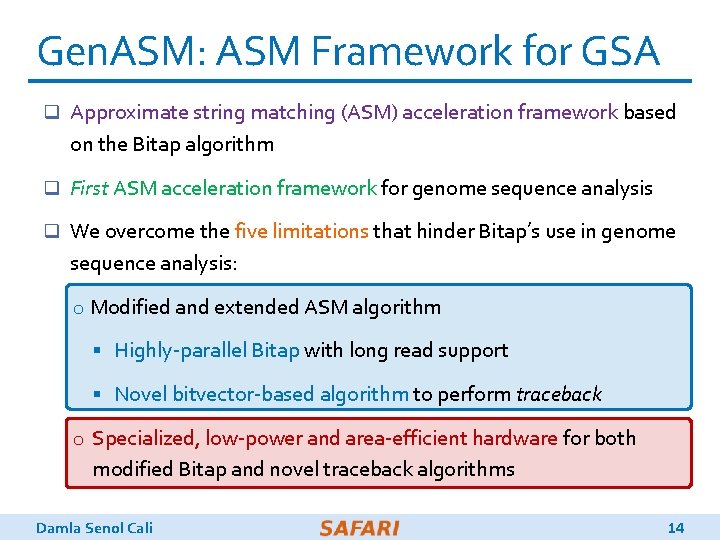
Gen. ASM: ASM Framework for GSA q Approximate string matching (ASM) acceleration framework based on the Bitap algorithm q First ASM acceleration framework for genome sequence analysis q We overcome the five limitations that hinder Bitap’s use in genome sequence analysis: o Modified and extended ASM algorithm § Highly-parallel Bitap with long read support § Novel bitvector-based algorithm to perform traceback o Specialized, low-power and area-efficient hardware for both modified Bitap and novel traceback algorithms Damla Senol Cali 14

Gen. ASM Algorithm q Gen. ASM-DC Algorithm: o Modified Bitap for Distance Calculation o Extended for efficient long read support o Besides bit-parallelism that Bitap has, extended for parallelism: § Loop unrolling § Text-level parallelism q Gen. ASM-TB Algorithm: o Novel Bitap-compatible Trace. Back algorithm o Walks through the intermediate bitvectors (match, deletion, substitution, insertion) generated by Gen. ASM-DC o Follows a divide-and-conquer approach to decrease the memory footprint Damla Senol Cali 15

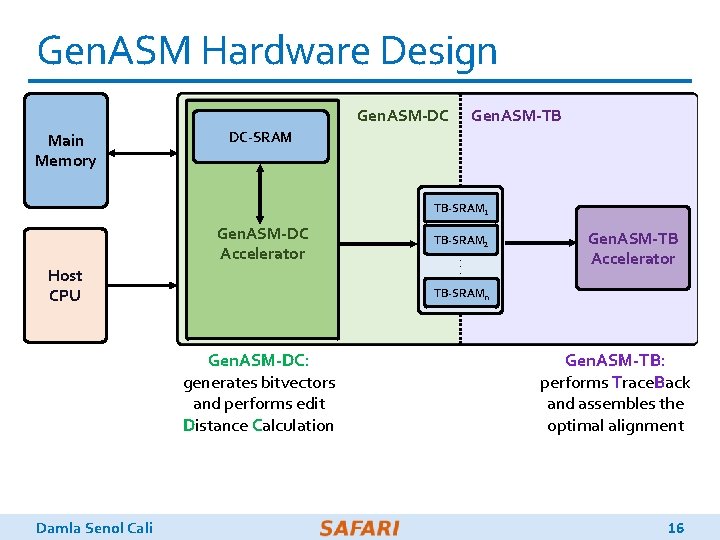
Gen. ASM Hardware Design Gen. ASM-DC Main Memory Gen. ASM-TB DC-SRAM TB-SRAM 1 Gen. ASM-DC Accelerator Host CPU . . . Gen. ASM-TB Accelerator TB-SRAMn Gen. ASM-DC: generates bitvectors and performs edit Distance Calculation Damla Senol Cali TB-SRAM 2 Gen. ASM-TB: performs Trace. Back and assembles the optimal alignment 16

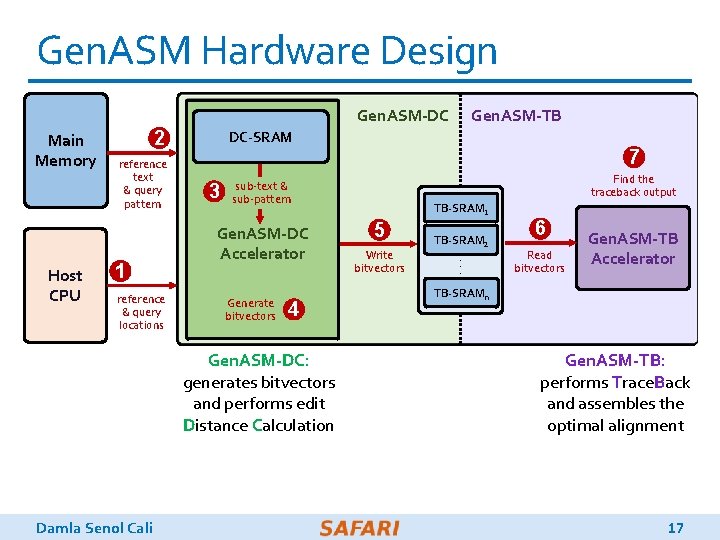
Gen. ASM Hardware Design Gen. ASM-DC Main Memory Host CPU 2 reference text & query pattern 1 reference & query locations DC-SRAM 3 7 Find the traceback output sub-text & sub-pattern Gen. ASM-DC Accelerator Generate bitvectors 4 Gen. ASM-DC: generates bitvectors and performs edit Distance Calculation Damla Senol Cali Gen. ASM-TB TB-SRAM 1 5 Write bitvectors TB-SRAM 2. . . 6 Read bitvectors Gen. ASM-TB Accelerator TB-SRAMn Gen. ASM-TB: performs Trace. Back and assembles the optimal alignment 17

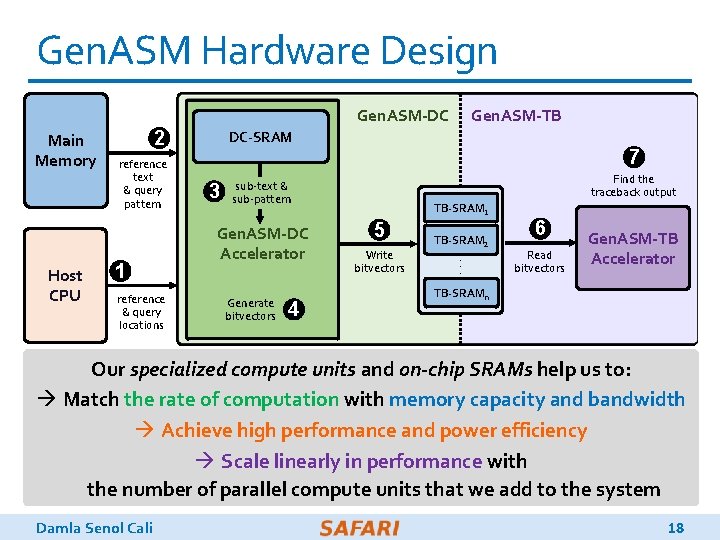
Gen. ASM Hardware Design Gen. ASM-DC Main Memory Host CPU 2 reference text & query pattern 1 reference & query locations Gen. ASM-TB DC-SRAM 3 7 Find the traceback output sub-text & sub-pattern Gen. ASM-DC Accelerator Generate bitvectors 4 TB-SRAM 1 5 Write bitvectors TB-SRAM 2. . . 6 Read bitvectors Gen. ASM-TB Accelerator TB-SRAMn Gen. ASM-DC: Gen. ASM-TB: Our specialized compute units and on-chip SRAMs help us to: generates bitvectors performs Trace. Back à Match the rateand performs edit of computation with memory capacity and assembles the and bandwidth Distance Calculation optimal alignment à Achieve high performance and power efficiency à Scale linearly in performance with the number of parallel compute units that we add to the system Damla Senol Cali 18

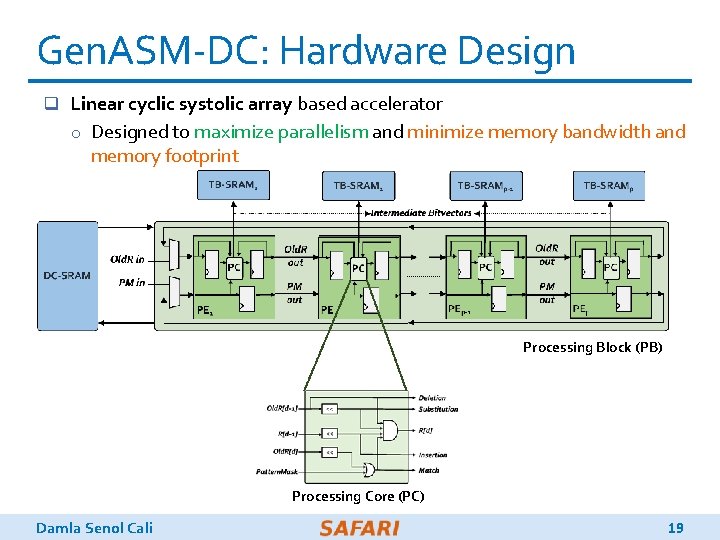
Gen. ASM-DC: Hardware Design q Linear cyclic systolic array based accelerator o Designed to maximize parallelism and minimize memory bandwidth and memory footprint Processing Block (PB) Processing Core (PC) Damla Senol Cali 19

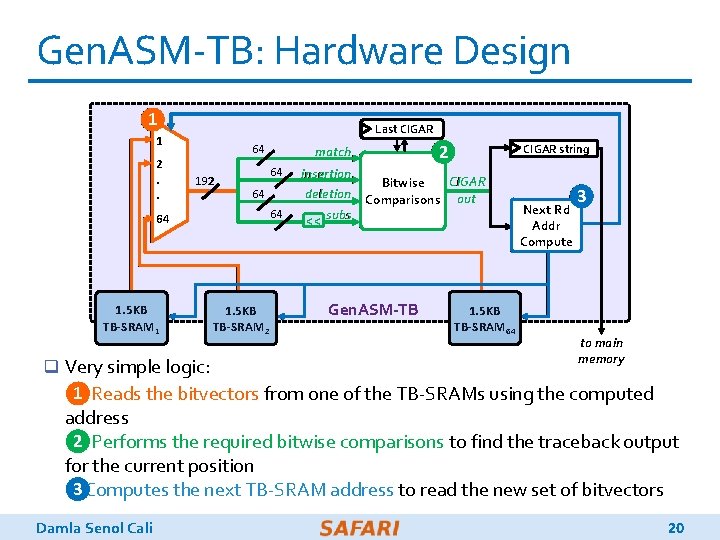
Gen. ASM-TB: Hardware Design 1 Last CIGAR 1 2. . 64 192 64 1. 5 KB TB-SRAM 1 64 1. 5 KB TB-SRAM 2 insertion deletion 64 64 2 match << subs CIGAR string CIGAR Bitwise Comparisons out Gen. ASM-TB 1. 5 KB TB-SRAM 64 Next Rd Addr Compute 3 to main memory q Very simple logic: 1 ❶Reads the bitvectors from one of the TB-SRAMs using the computed address 2 ❷Performs the required bitwise comparisons to find the traceback output for the current position 3 ❸Computes the next TB-SRAM address to read the new set of bitvectors Damla Senol Cali 20

Use Cases of Gen. ASM (1) Read Alignment Step of Read Mapping o Find the optimal alignment of how reads map to candidate reference regions (2) Pre-Alignment Filtering for Short Reads o Quickly identify and filter out the unlikely candidate reference regions for each read (3) Edit Distance Calculation o Measure the similarity or distance between two sequences q We also discuss other possible use cases of Gen. ASM in our paper: o Read-to-read overlap finding, hash-table based indexing, whole genome alignment, generic text search Damla Senol Cali 21

Outline q Introduction q Background o Approximate String Matching (ASM) o ASM with Bitap Algorithm q Gen. ASM: ASM Acceleration Framework o Gen. ASM Algorithm o Gen. ASM Hardware Design o Use Cases of Gen. ASM q Evaluation q Conclusion Damla Senol Cali 22

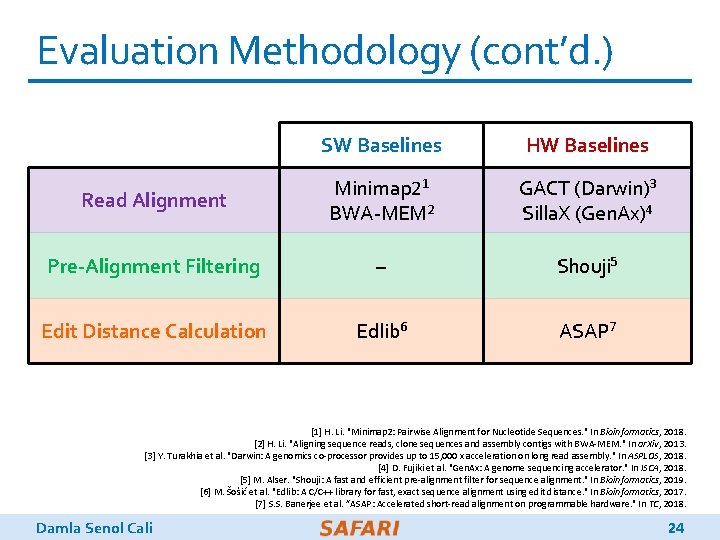
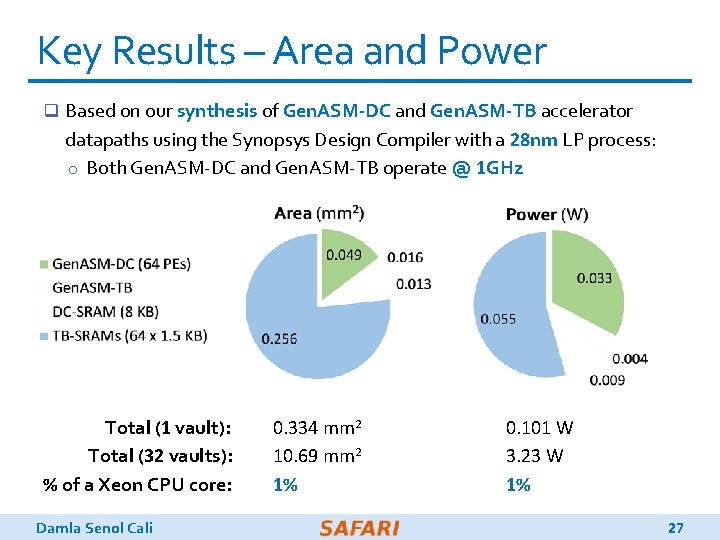
Evaluation Methodology q We evaluate Gen. ASM using: o Synthesized System. Verilog models of the Gen. ASM-DC and Gen. ASM-TB accelerator datapaths o Detailed simulation-based performance modeling q 16 GB HMC-like 3 D-stacked DRAM architecture o 32 vaults o 256 GB/s of internal bandwidth, clock frequency of 1. 25 GHz o In order to achieve high parallelism and low power-consumption o Within each vault, the logic layer contains a Gen. ASM-DC accelerator, its associated DC-SRAM, a Gen. ASM-TB accelerator, and TB-SRAMs. Damla Senol Cali 23

Evaluation Methodology (cont’d. ) SW Baselines HW Baselines Read Alignment Minimap 21 BWA-MEM 2 GACT (Darwin)3 Silla. X (Gen. Ax)4 Pre-Alignment Filtering – Shouji 5 Edit Distance Calculation Edlib 6 ASAP 7 [1] H. Li. "Minimap 2: Pairwise Alignment for Nucleotide Sequences. " In Bioinformatics, 2018. [2] H. Li. "Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. " In ar. Xiv, 2013. [3] Y. Turakhia et al. "Darwin: A genomics co-processor provides up to 15, 000 x acceleration on long read assembly. " In ASPLOS, 2018. [4] D. Fujiki et al. "Gen. Ax: A genome sequencing accelerator. " In ISCA, 2018. [5] M. Alser. "Shouji: A fast and efficient pre-alignment filter for sequence alignment. " In Bioinformatics, 2019. [6] M. Šošić et al. "Edlib: A C/C++ library for fast, exact sequence alignment using edit distance. " In Bioinformatics, 2017. [7] S. S. Banerjee et al. ”ASAP: Accelerated short-read alignment on programmable hardware. " In TC, 2018. Damla Senol Cali 24

Evaluation Methodology (cont’d. ) q For Use Case 1: Read Alignment, we compare Gen. ASM with: o Minimap 2 and BWA-MEM (state-of-the-art SW) § Running on Intel® Xeon® Gold 6126 CPU (12 -core) operating @2. 60 GHz with 64 GB DDR 4 memory § Using two simulated datasets: ◦ Long ONT and Pac. Bio reads: 10 Kbp reads, 10 -15% error rate ◦ Short Illumina reads: 100 -250 bp reads, 5% error rate o GACT of Darwin and Silla. X of Gen. Ax (state-of-the-art HW) § Open-source RTL for GACT § Data reported by the original work for Silla. X § GACT is best for long reads, Silla. X is best for short reads Damla Senol Cali 25

Evaluation Methodology (cont’d. ) q For Use Case 2: Pre-Alignment Filtering, we compare Gen. ASM with: o Shouji (state-of-the-art HW – FPGA-based filter) § Using two datasets provided as test cases: • 100 bp reference-read pairs with an edit distance threshold of 5 • 250 bp reference-read pairs with an edit distance threshold of 15 q For Use Case 3: Edit Distance Calculation, we compare Gen. ASM with: o Edlib (state-of-the-art SW) § Using two 100 Kbp and 1 Mbp sequences with similarity ranging between 60%-99% o ASAP (state-of-the-art HW – FPGA-based accelerator) § Using data reported by the original work Damla Senol Cali 26

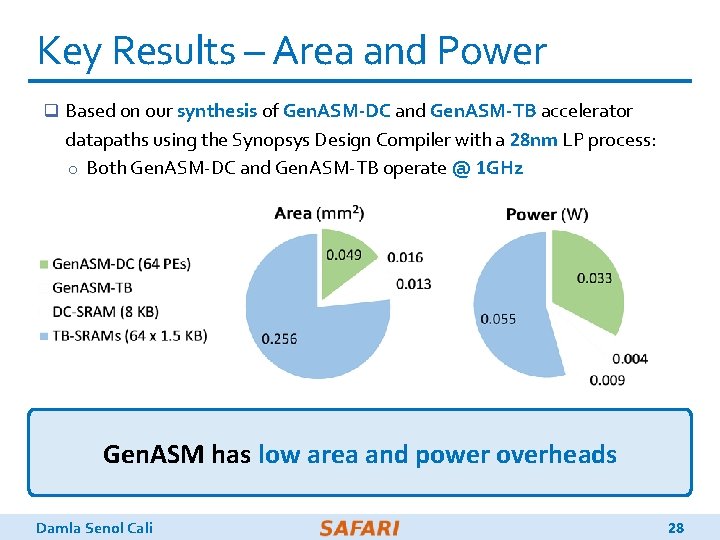
Key Results – Area and Power q Based on our synthesis of Gen. ASM-DC and Gen. ASM-TB accelerator datapaths using the Synopsys Design Compiler with a 28 nm LP process: o Both Gen. ASM-DC and Gen. ASM-TB operate @ 1 GHz Total (1 vault): 0. 334 mm 2 Total (32 vaults): 10. 69 mm 2 % of a Xeon CPU core: 1% Damla Senol Cali 0. 101 W 3. 23 W 1% 27

Key Results – Area and Power q Based on our synthesis of Gen. ASM-DC and Gen. ASM-TB accelerator datapaths using the Synopsys Design Compiler with a 28 nm LP process: o Both Gen. ASM-DC and Gen. ASM-TB operate @ 1 GHz Gen. ASM has low area and power overheads Damla Senol Cali 28

Key Results – Use Case 1 (1) Read Alignment Step of Read Mapping o Find the optimal alignment of how reads map to candidate reference regions (2) Pre-Alignment Filtering for Short Reads o Quickly identify and filter out the unlikely candidate reference regions for each read (3) Edit Distance Calculation o Measure the similarity or distance between two sequences Damla Senol Cali 29

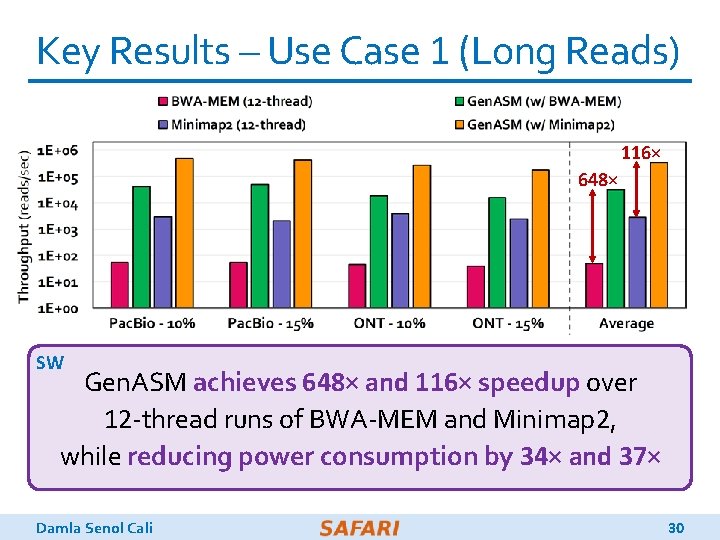
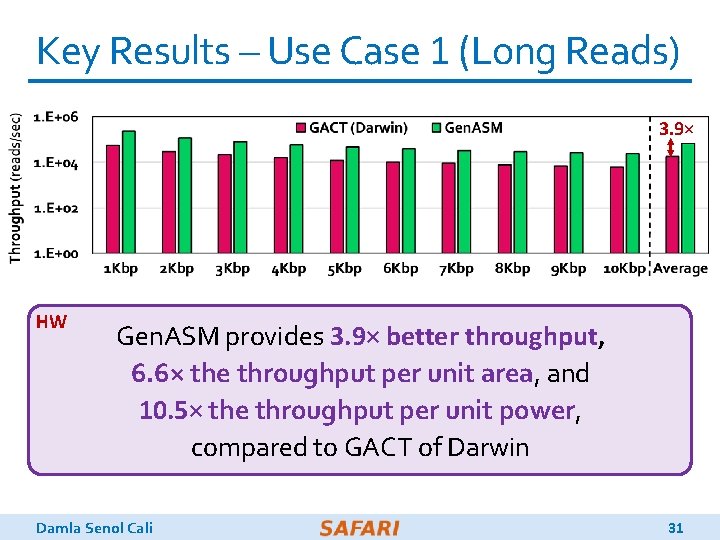
Key Results – Use Case 1 (Long Reads) 116× 648× SW Gen. ASM achieves 648× and 116× speedup over 12 -thread runs of BWA-MEM and Minimap 2, while reducing power consumption by 34× and 37× Damla Senol Cali 30

Key Results – Use Case 1 (Long Reads) 3. 9× HW Gen. ASM provides 3. 9× better throughput, 6. 6× the throughput per unit area, and 10. 5× the throughput per unit power, compared to GACT of Darwin Damla Senol Cali 31

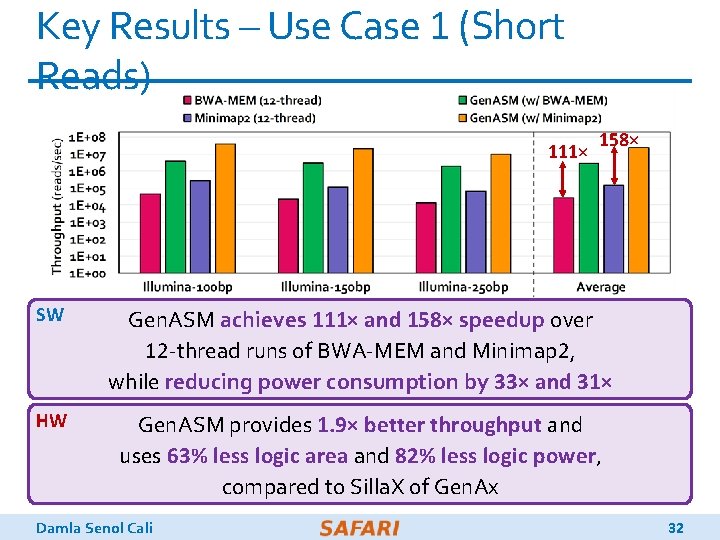
Key Results – Use Case 1 (Short Reads) 111× 158× SW Gen. ASM achieves 111× and 158× speedup over 12 -thread runs of BWA-MEM and Minimap 2, while reducing power consumption by 33× and 31× HW Gen. ASM provides 1. 9× better throughput and uses 63% less logic area and 82% less logic power, compared to Silla. X of Gen. Ax Damla Senol Cali 32

Key Results – Use Case 2 (1) Read Alignment Step of Read Mapping o Find the optimal alignment of how reads map to candidate reference regions (2) Pre-Alignment Filtering for Short Reads o Quickly identify and filter out the unlikely candidate reference regions for each read (3) Edit Distance Calculation o Measure the similarity or distance between two sequences Damla Senol Cali 33

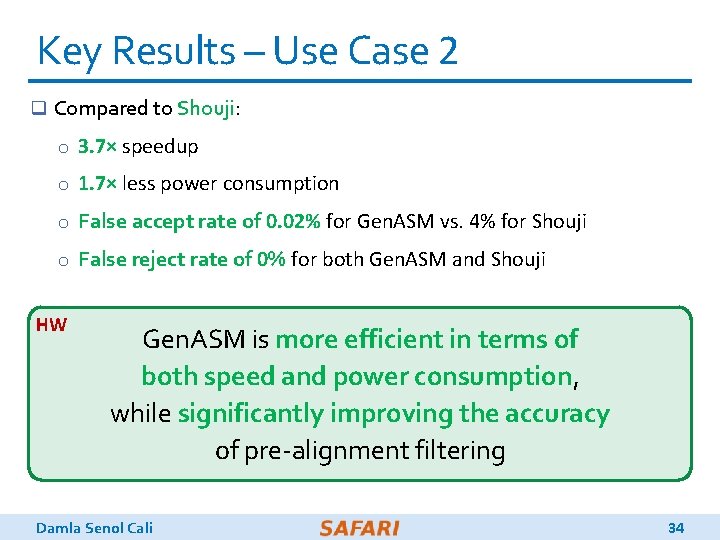
Key Results – Use Case 2 q Compared to Shouji: o 3. 7× speedup o 1. 7× less power consumption o False accept rate of 0. 02% for Gen. ASM vs. 4% for Shouji o False reject rate of 0% for both Gen. ASM and Shouji HW Gen. ASM is more efficient in terms of both speed and power consumption, while significantly improving the accuracy of pre-alignment filtering Damla Senol Cali 34

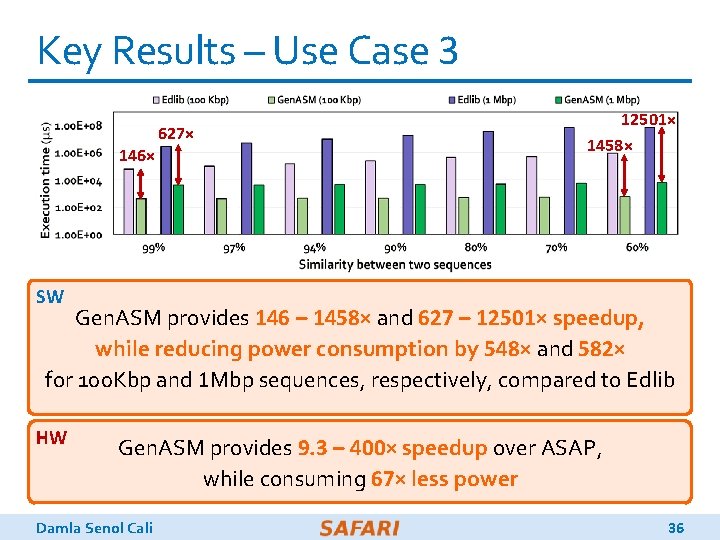
Key Results – Use Case 3 (1) Read Alignment Step of Read Mapping o Find the optimal alignment of how reads map to candidate reference regions (2) Pre-Alignment Filtering for Short Reads o Quickly identify and filter out the unlikely candidate reference regions for each read (3) Edit Distance Calculation o Measure the similarity or distance between two sequences Damla Senol Cali 35

Key Results – Use Case 3 146× 627× 12501× 1458× SW Gen. ASM provides 146 – 1458× and 627 – 12501× speedup, while reducing power consumption by 548× and 582× for 100 Kbp and 1 Mbp sequences, respectively, compared to Edlib HW Gen. ASM provides 9. 3 – 400× speedup over ASAP, while consuming 67× less power Damla Senol Cali 36

Outline q Introduction q Background o Approximate String Matching (ASM) o ASM with Bitap Algorithm q Gen. ASM: ASM Acceleration Framework o Gen. ASM Algorithm o Gen. ASM Hardware Design o Use Cases of Gen. ASM q Evaluation q Conclusion Damla Senol Cali 37

Additional Details in the Paper q Details of the Gen. ASM-DC and Gen. ASM-TB algorithms q Big-O analysis of the algorithms q Detailed explanation of evaluated use cases q Evaluation methodology details (datasets, baselines, performance model) q Additional results for the three evaluated use cases q Sources of improvements in Gen. ASM (algorithm-level, hardware-level, technology-level) q Discussion of four other potential use cases of Gen. ASM Damla Senol Cali 38

Conclusion q Problem: o Genome sequence analysis is bottlenecked by the computational power and memory bandwidth limitations of existing systems o This bottleneck is particularly an issue for approximate string matching q Key Contributions: o Gen. ASM: An approximate string matching (ASM) acceleration framework to accelerate multiple steps of genome sequence analysis § First to enhance and accelerate Bitap for ASM with genomic sequences § Co-design of our modified scalable and memory-efficient algorithms with low-power and area-efficient hardware accelerators § Evaluation of three different use cases: read alignment, pre-alignment filtering, edit distance calculation q Key Results: Gen. ASM is significantly more efficient for all the three use cases (in terms of throughput and throughput per unit power) than state-of-the-art software and hardware baselines Damla Senol Cali 39

Gen. ASM: A High Performance, Low-Power Approximate String Matching Acceleration Framework for Genome Sequence Analysis Damla Senol Cali Carnegie Mellon University (dsenol@andrew. cmu. edu) Gurpreet S. Kalsi 2, Zulal Bingol 3, Can Firtina 4, Lavanya Subramanian 5, Jeremie Kim 1, 4, Rachata Ausavarungnirun 6, 1, Mohammed Alser 4, Juan Gomez-Luna 4, Amirali Boroumand 1, Anant Nori 2, Allison Scibisz 1, Sreenivas Subramoney 2, Can Alkan 3, Saugata Ghose 7, 1, and Onur Mutlu 4, 1, 3 1 2 5 6 3 4 7 1, 4

Backup Slides

Genome Sequencing Large DNA molecule Small DNA fragments ACGTACCCCGT TTTTTTTAATT AAAAA GATACACTGT G ACGACGTAGCT CTAGGGACCTT ACGAGCGGGT Damla Senol Cali Reads 42

Read Mapping Reference genome Indexing Hash-table based index Reads Reference segment Seeding Potential mapping locations Pre-Alignment Filtering Query read Remaining potential mapping locations Read Alignment Damla Senol Cali Optimal alignment 43

Short Reads vs. Long Reads Ø Short Reads q Sequences with tens to hundreds of bases q Highly accurate sequences q Output of SRS technologies (e. g. , Illumina, Ion Torrent) Ø Long reads q Sequences with thousands or millions of bases q Sequences with high error rates q Output of LRS technologies (e. g. , Oxford Nanopore Technologies, Pac. Bio) Damla Senol Cali 44

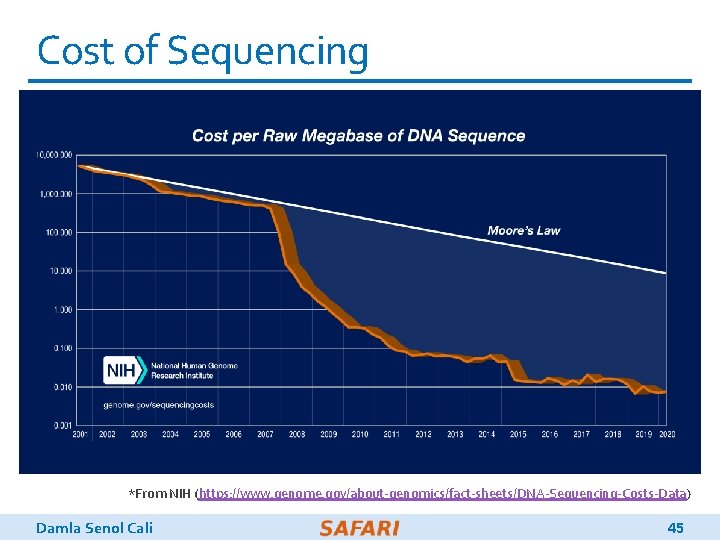
Cost of Sequencing *From NIH (https: //www. genome. gov/about-genomics/fact-sheets/DNA-Sequencing-Costs-Data) Damla Senol Cali 45

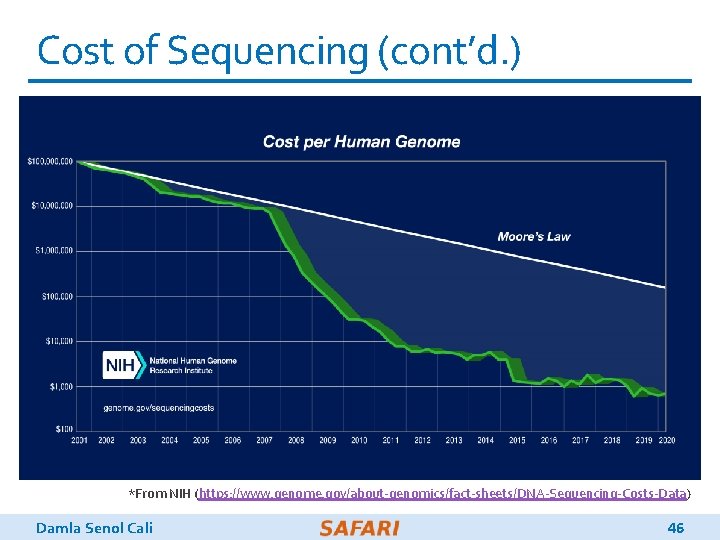
Cost of Sequencing (cont’d. ) *From NIH (https: //www. genome. gov/about-genomics/fact-sheets/DNA-Sequencing-Costs-Data) Damla Senol Cali 46

Sequencing of COVID-19 q Why whole genome sequencing (WGS) and sequence data analysis are important: o To detect the virus from a human sample such as saliva, Bronchoalveolar fluid etc. o To understand the sources and modes of transmission of the virus o To discover the genomic characteristics of the virus, and compare with the previous viruses (e. g. , 02 -03 SARS epidemic) o To design and evaluate the diagnostic tests q Two key areas of COVID-19 genomic research: o To sequence the genome of the virus itself, COVID-19, in order to track the mutations in the virus. o To explore the genes of infected patients. This analysis can be used to understand why some people get more severe symptoms than others, as well as, help with the development of new treatments in the future. Damla Senol Cali 47

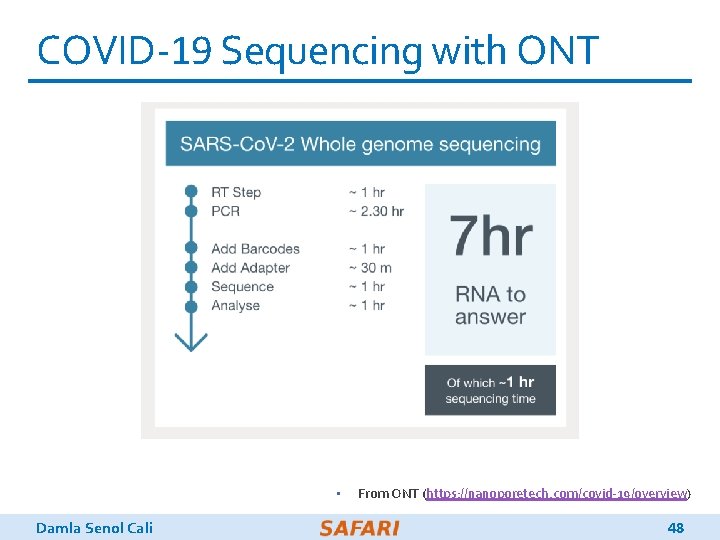
COVID-19 Sequencing with ONT • Damla Senol Cali From ONT (https: //nanoporetech. com/covid-19/overview) 48

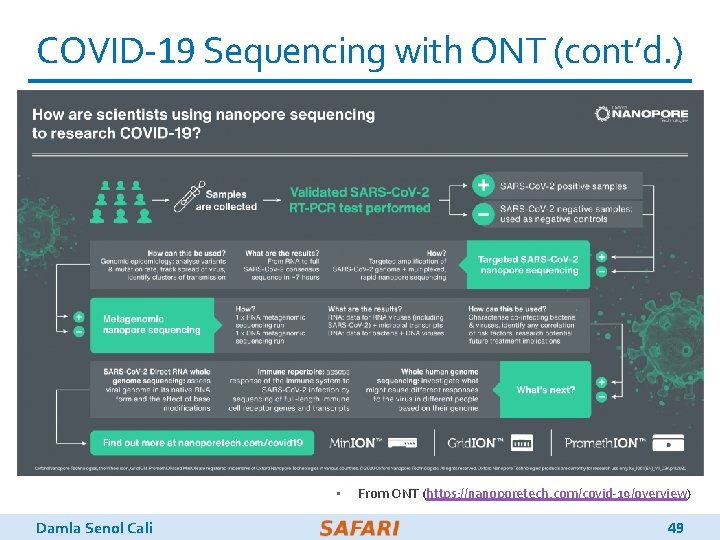
COVID-19 Sequencing with ONT (cont’d. ) • Damla Senol Cali From ONT (https: //nanoporetech. com/covid-19/overview) 49

Future of Genome Sequencing & Analysis Min. ION from ONT Smidg. ION from ONT Damla Senol Cali 50

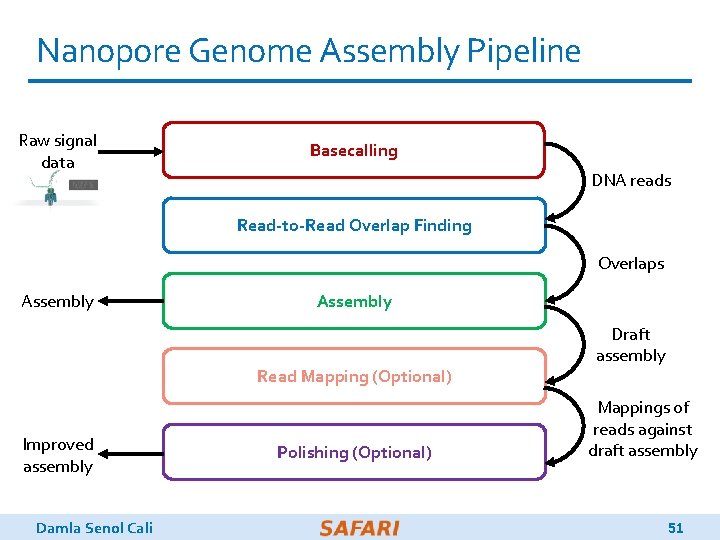
Nanopore Genome Assembly Pipeline Raw signal data Basecalling DNA reads Read-to-Read Overlap Finding Overlaps Assembly Read Mapping (Optional) Improved assembly Damla Senol Cali Polishing (Optional) Draft assembly Mappings of reads against draft assembly 51

Nanopore Sequencing & Tools Damla Senol Cali, Jeremie S. Kim, Saugata Ghose, Can Alkan, and Onur Mutlu. "Nanopore Sequencing Technology and Tools for Genome Assembly: Computational Analysis of the Current State, Bottlenecks and Future Directions. " Briefings in Bioinformatics (2018). Damla Senol Cali Bi. B Version ar. Xiv Version 52