GCP 21 II S 3 Cassava Genome from















- Slides: 38

GCP 21 -II S 3 Cassava Genome from Ancestor to Cultivar Wenquan Wang Ph. D Chinese Cassava Genomics Consortium Institute of Tropical Biosciences & Biotechnology, CATAS Uganda, June 19, 2012

Biological characteristics of cassava • High photosynthesis • High starch accumulation • Extremely tolerance to drought and barren soil • Heterozygosity and somatic propagation.

Bottleneck in aspect of genetics for developing cassava industry l l l Less known genetic diversity in evolution Lack knowledge for mechanisms of high photosynthesis and starch metabolism Uncovering function of drought and barren soil tolerance Less understanding adaptation to different kinds of diseases and pests of cassava plant Lack tools for genotyping in cassava breeding

Genotypes used for whole genome sequencing W 14 (Manihot esculenta. ssp. flabellifolia) Semi-wild species KU 50 (Manihot esculanta Crantz) Cultivar (starchy) S 1. 600 (Manihot esculanta Crantz) Cultivar (sugary ) KU 50 W 14

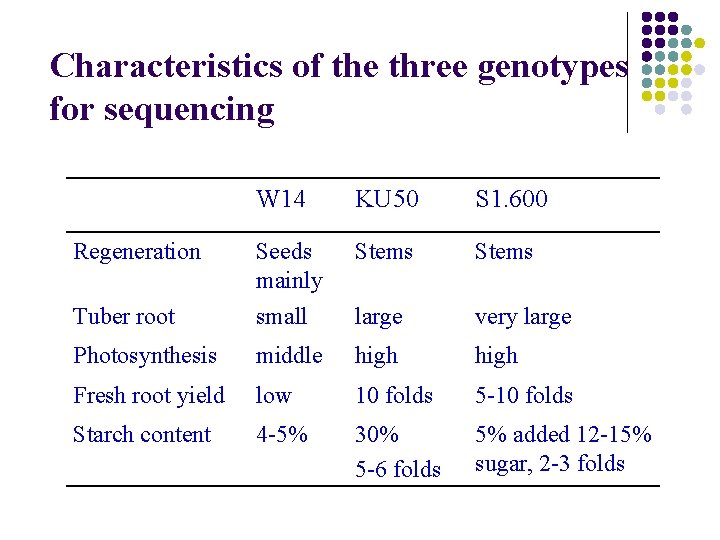
Characteristics of the three genotypes for sequencing W 14 KU 50 S 1. 600 Stems Tuber root Seeds mainly small large very large Photosynthesis middle high Fresh root yield low 10 folds 5 -10 folds Starch content 4 -5% 30% 5 -6 folds 5% added 12 -15% sugar, 2 -3 folds Regeneration

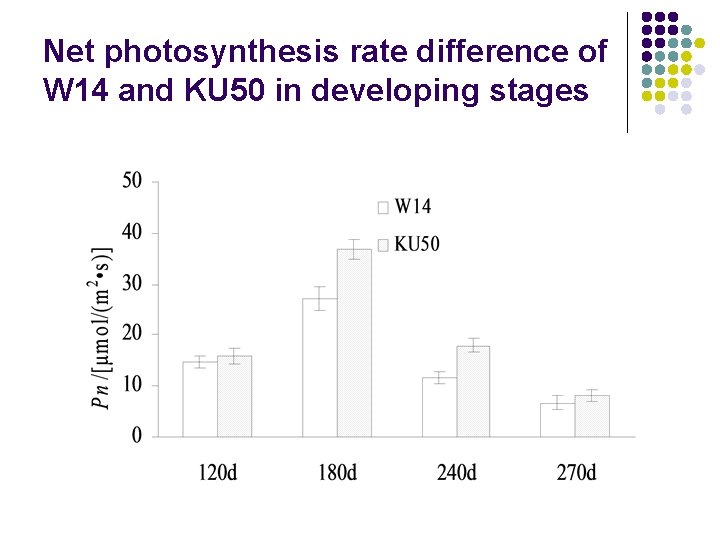
Net photosynthesis rate difference of W 14 and KU 50 in developing stages

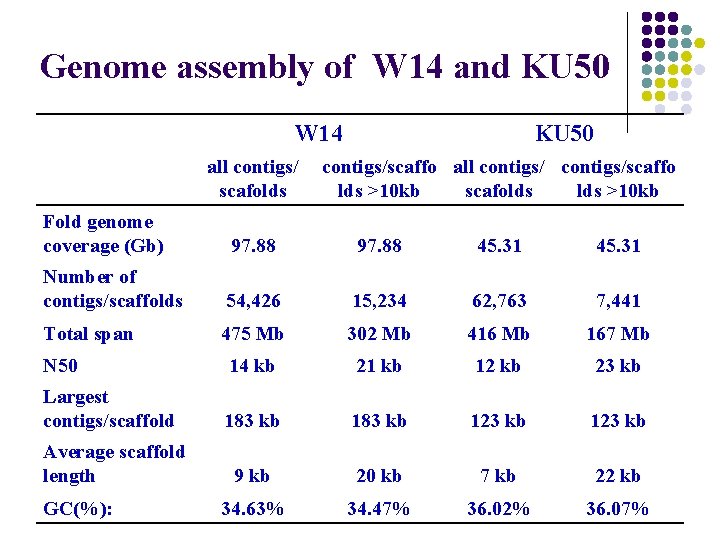
Genome assembly of W 14 and KU 50 W 14 all contigs/ scafolds KU 50 contigs/scaffo all contigs/scaffo lds >10 kb scafolds >10 kb Fold genome coverage (Gb) 97. 88 45. 31 Number of contigs/scaffolds 54, 426 15, 234 62, 763 7, 441 Total span 475 Mb 302 Mb 416 Mb 167 Mb N 50 14 kb 21 kb 12 kb 23 kb Largest contigs/scaffold 183 kb 123 kb 9 kb 20 kb 7 kb 22 kb 34. 63% 34. 47% 36. 02% 36. 07% Average scaffold length GC(%):

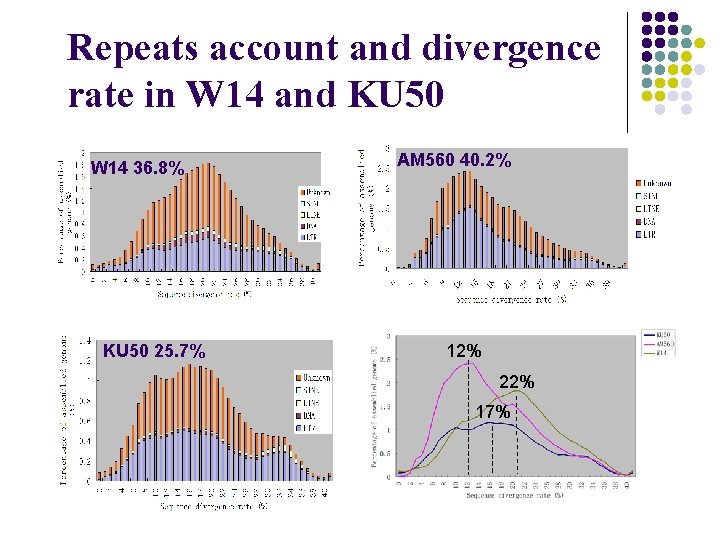
Repeats account and divergence rate in W 14 and KU 50 W 14 36. 8% KU 50 25. 7% AM 560 40. 2% 12% 22% 17%

LTR in situ hybridization in all the chromosomes a b

Genome coverage in gene region W 14 Transcripts coverage 97. 1% W 14 EST coverage 91. 5% KU 50 Transcript coverage 80. 5% KU 50 EST coverage 73. 4%

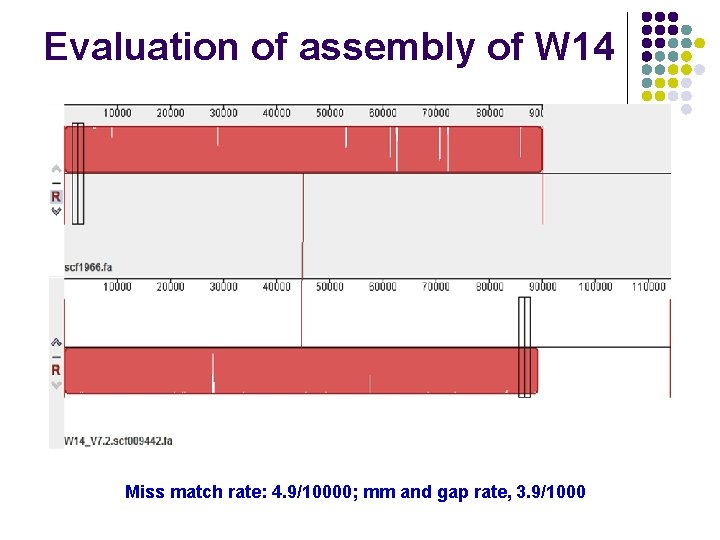
Evaluation of assembly of W 14 Miss match rate: 4. 9/10000; mm and gap rate, 3. 9/1000

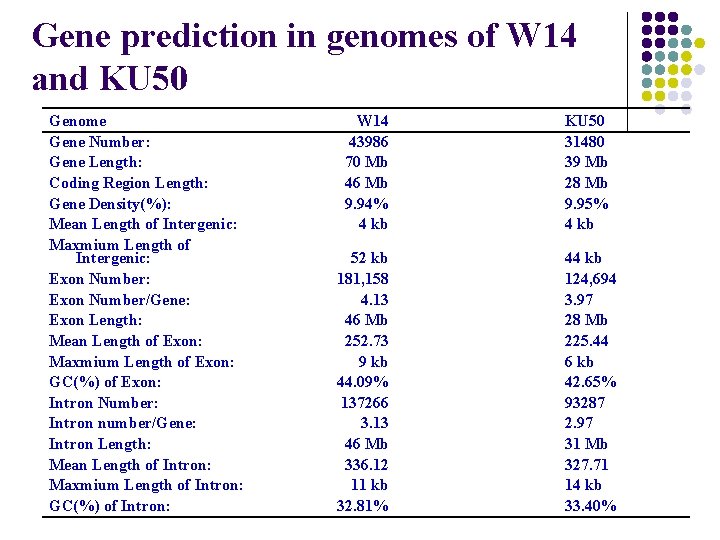
Gene prediction in genomes of W 14 and KU 50 Genome Gene Number: Gene Length: Coding Region Length: Gene Density(%): Mean Length of Intergenic: Maxmium Length of Intergenic: Exon Number/Gene: Exon Length: Mean Length of Exon: Maxmium Length of Exon: GC(%) of Exon: Intron Number: Intron number/Gene: Intron Length: Mean Length of Intron: Maxmium Length of Intron: GC(%) of Intron: W 14 43986 70 Mb 46 Mb 9. 94% 4 kb KU 50 31480 39 Mb 28 Mb 9. 95% 4 kb 52 kb 181, 158 4. 13 46 Mb 252. 73 9 kb 44. 09% 137266 3. 13 46 Mb 336. 12 11 kb 32. 81% 44 kb 124, 694 3. 97 28 Mb 225. 44 6 kb 42. 65% 93287 2. 97 31 Mb 327. 71 14 kb 33. 40%

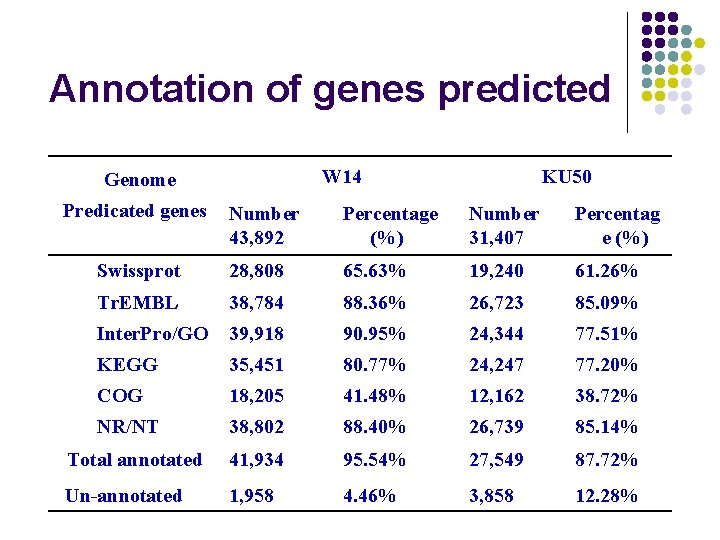
Annotation of genes predicted W 14 Genome Predicated genes KU 50 Number 43, 892 Percentage (%) Number 31, 407 Percentag e (%) Swissprot 28, 808 65. 63% 19, 240 61. 26% Tr. EMBL 38, 784 88. 36% 26, 723 85. 09% Inter. Pro/GO 39, 918 90. 95% 24, 344 77. 51% KEGG 35, 451 80. 77% 24, 247 77. 20% COG 18, 205 41. 48% 12, 162 38. 72% NR/NT 38, 802 88. 40% 26, 739 85. 14% 41, 934 95. 54% 27, 549 87. 72% 1, 958 4. 46% 3, 858 12. 28% Total annotated Un-annotated

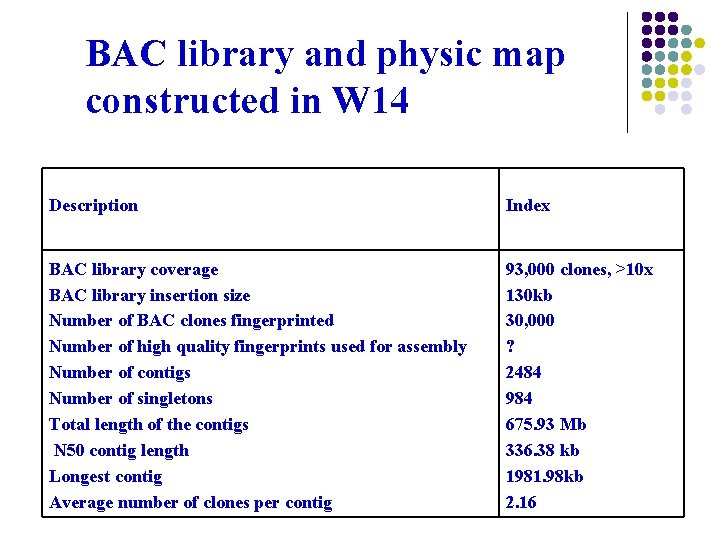
BAC library and physic map constructed in W 14 Description Index BAC library coverage BAC library insertion size Number of BAC clones fingerprinted Number of high quality fingerprints used for assembly Number of contigs Number of singletons Total length of the contigs N 50 contig length Longest contig Average number of clones per contig 93, 000 clones, >10 x 130 kb 30, 000 ? 2484 984 675. 93 Mb 336. 38 kb 1981. 98 kb 2. 16

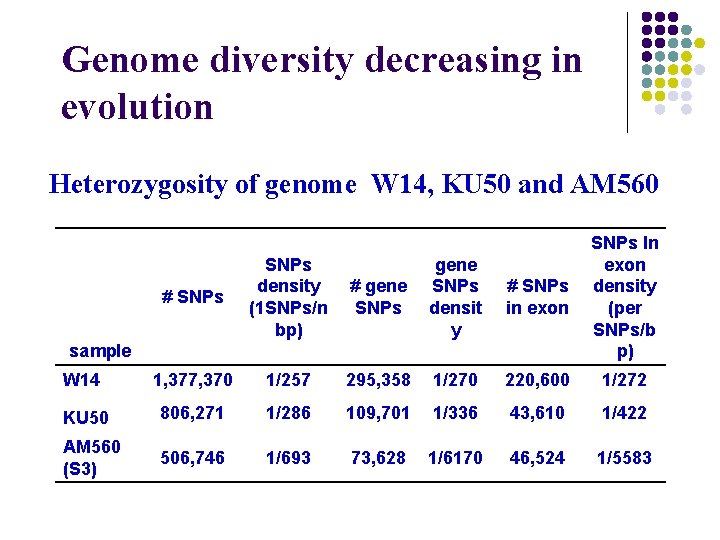
Genome diversity decreasing in evolution Heterozygosity of genome W 14, KU 50 and AM 560 # gene SNPs densit y # SNPs in exon SNPs In exon density (per SNPs/b p) 1/257 295, 358 1/270 220, 600 1/272 806, 271 1/286 109, 701 1/336 43, 610 1/422 506, 746 1/693 73, 628 1/6170 46, 524 1/5583 # SNPs density (1 SNPs/n bp) W 14 1, 377, 370 KU 50 AM 560 (S 3) sample

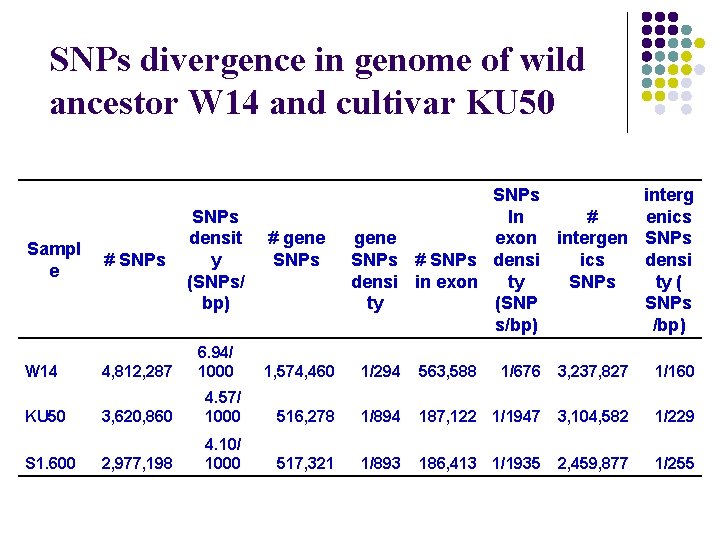
SNPs divergence in genome of wild ancestor W 14 and cultivar KU 50 Sampl e W 14 KU 50 S 1. 600 # SNPs 4, 812, 287 SNPs densit y (SNPs/ bp) 6. 94/ 1000 # gene SNPs interg # In enics gene exon intergen SNPs ics SNPs # SNPs densi in exon SNPs ty ty (SNP SNPs s/bp) 1, 574, 460 1/294 563, 588 1/676 3, 237, 827 1/160 3, 620, 860 4. 57/ 1000 516, 278 1/894 187, 122 1/1947 3, 104, 582 1/229 2, 977, 198 4. 10/ 1000 517, 321 1/893 186, 413 1/1935 2, 459, 877 1/255

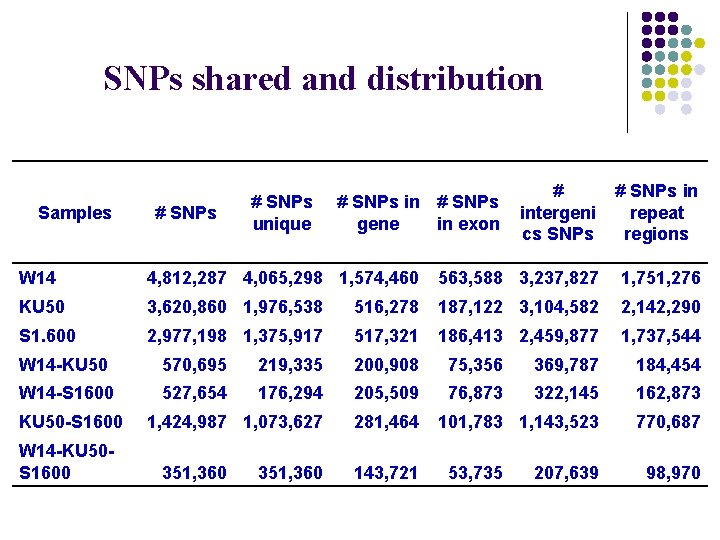
SNPs shared and distribution Samples # SNPs unique # SNPs in # SNPs gene in exon # intergeni cs SNPs # SNPs in repeat regions W 14 4, 812, 287 4, 065, 298 1, 574, 460 563, 588 3, 237, 827 1, 751, 276 KU 50 3, 620, 860 1, 976, 538 516, 278 187, 122 3, 104, 582 2, 142, 290 S 1. 600 2, 977, 198 1, 375, 917 517, 321 186, 413 2, 459, 877 1, 737, 544 W 14 -KU 50 570, 695 219, 335 200, 908 75, 356 369, 787 184, 454 W 14 -S 1600 527, 654 176, 294 205, 509 76, 873 322, 145 162, 873 1, 424, 987 1, 073, 627 281, 464 101, 783 1, 143, 523 770, 687 KU 50 -S 1600 W 14 -KU 50 S 1600 351, 360 143, 721 53, 735 207, 639 98, 970

Indels divergence in genome of wild ancestor W 14 and cultivar KU 50 Sample # indels density # insertion # deletion average length W 14 390, 652 0. 80/1000 159, 467 231, 080 3. 59 KU 50 275, 639 0. 79/1000 132, 396 143, 200 3. 65 S 1. 600 217, 226 0. 64/1000 103, 964 113, 207 4. 07

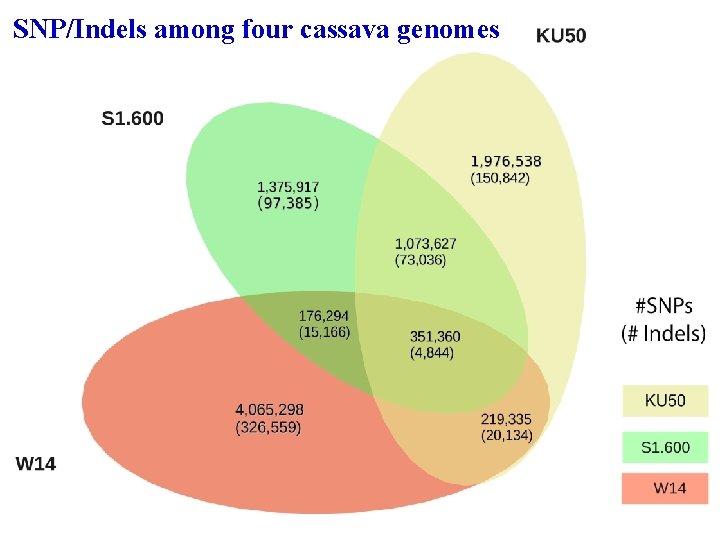
SNP/Indels among four cassava genomes

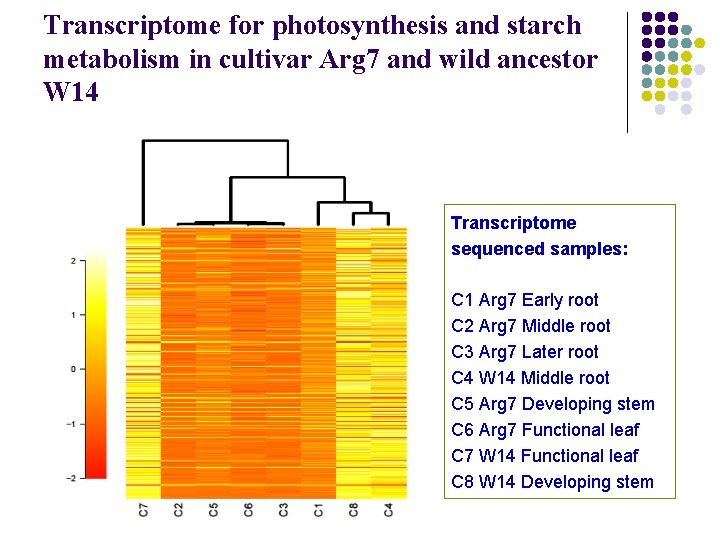
Transcriptome for photosynthesis and starch metabolism in cultivar Arg 7 and wild ancestor W 14 Transcriptome sequenced samples: C 1 Arg 7 Early root C 2 Arg 7 Middle root C 3 Arg 7 Later root C 4 W 14 Middle root C 5 Arg 7 Developing stem C 6 Arg 7 Functional leaf C 7 W 14 Functional leaf C 8 W 14 Developing stem

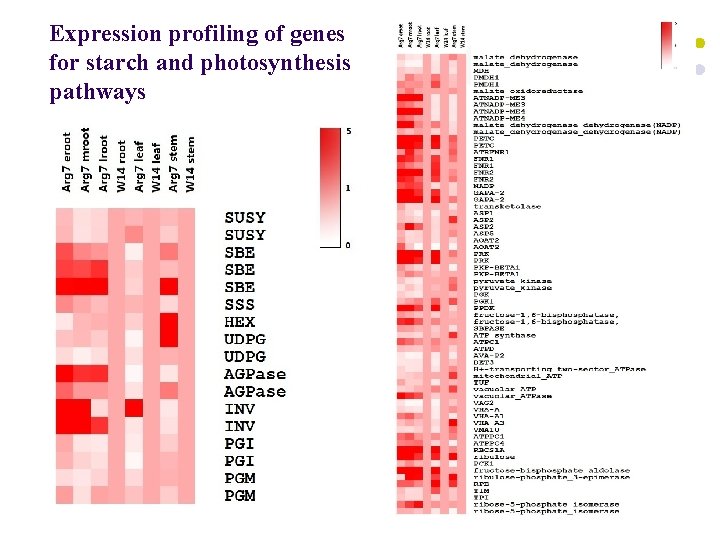
Expression profiling of genes for starch and photosynthesis pathways

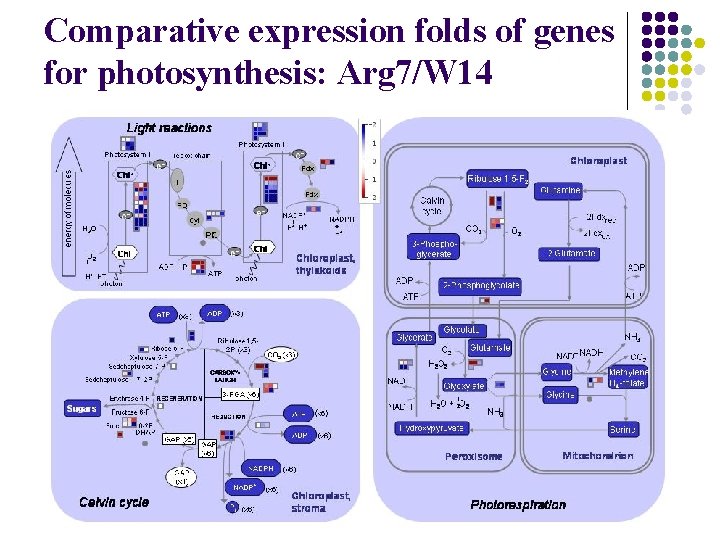
Comparative expression folds of genes for photosynthesis: Arg 7/W 14

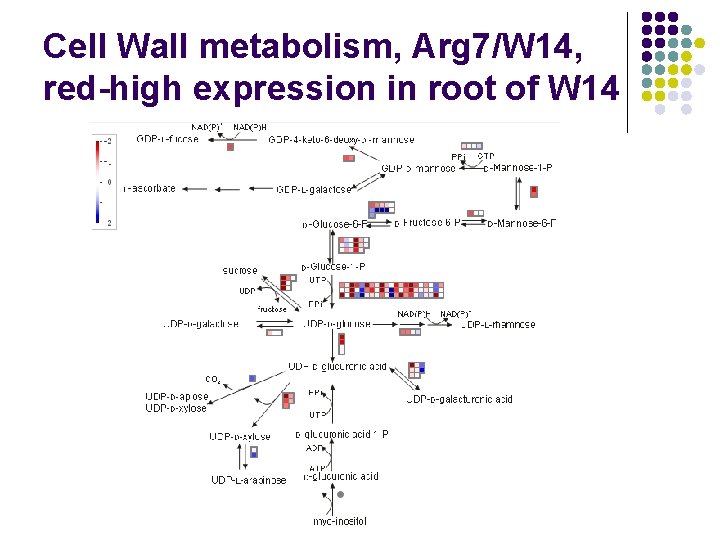
Cell Wall metabolism, Arg 7/W 14, red-high expression in root of W 14

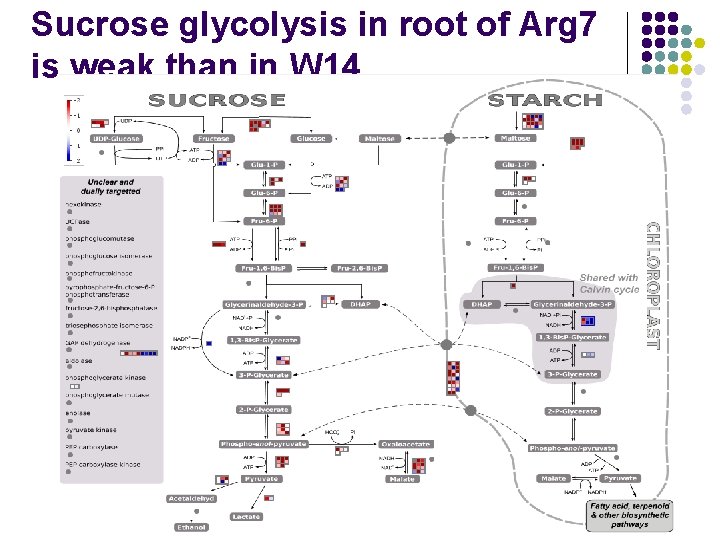
Sucrose glycolysis in root of Arg 7 is weak than in W 14

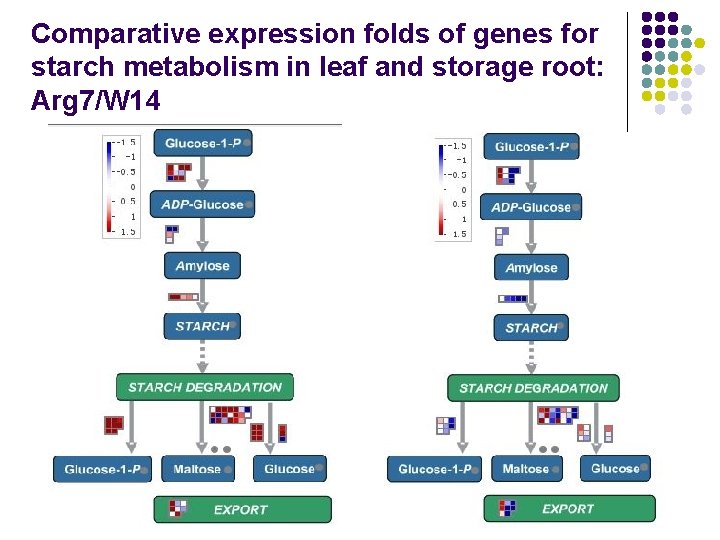
Comparative expression folds of genes for starch metabolism in leaf and storage root: Arg 7/W 14

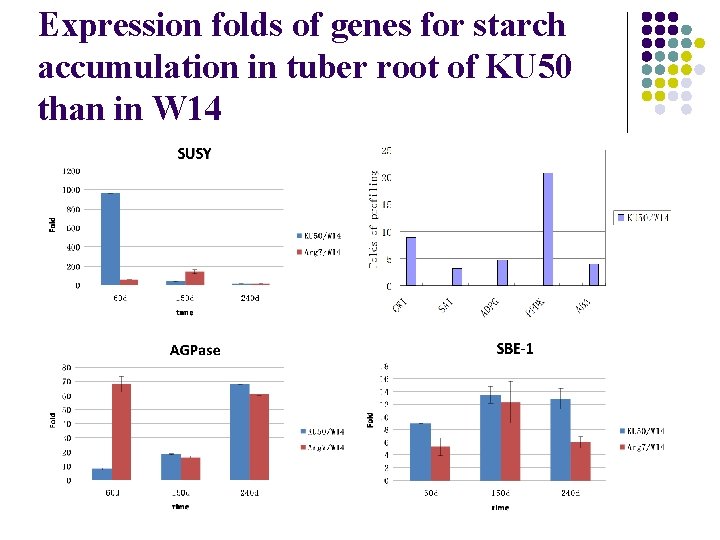
Expression folds of genes for starch accumulation in tuber root of KU 50 than in W 14

Phylogenetic tree of Su. Sy and INV Su. Sy INV

An efficient starch biosynthesis model in tuber root of cassava

mi. RNAs and drought tolerance in cassava l l a set of 148 mi. RNAs in cassava have been predicted by sequencing of 14 small RNA samples and referenced to genome which of 41 are novels and 107 are conserved. mi. RNAs and targets related to drought and development of leaf and tuber root have been found.

Eleven drought and cold inducible mi. RNAs with interesting targets were revealed and confirmed by q. PCR mi. RNA Targets mi. R 1045125 Protein binding, Zinc ion binding; Transcription factor, DNA binding mi. R 1230481 zinc ion binding, protein serine/threonine kinase, ATP binding mi. R 3747522 Enzyme inhibitor activity, Pectinesterase activity; DNA-binding protein -related; FUNCTIONS IN: transcription factor activity mi. R 5178028 Encodes a H 3/H 4 histone acetyltransferase; Encodes eukaryotic translation initiation factor. mi. R 3615546 oxidative phosphorylation uncoupler activity, binding oxidoreductase activity, Iron ion binding mi. R 1229496 Protein kinase family--kinase activity, small molecular g-protein mi. R 4806982 Kinase activity; Nuclear protein required for early embryogenesis auxin induced gene (IAA 1) encoding a short-lived nuclear-localized mi. R 5815094 transcriptional regulator protein; acetylglucosaminyltransferase ; transferase activity

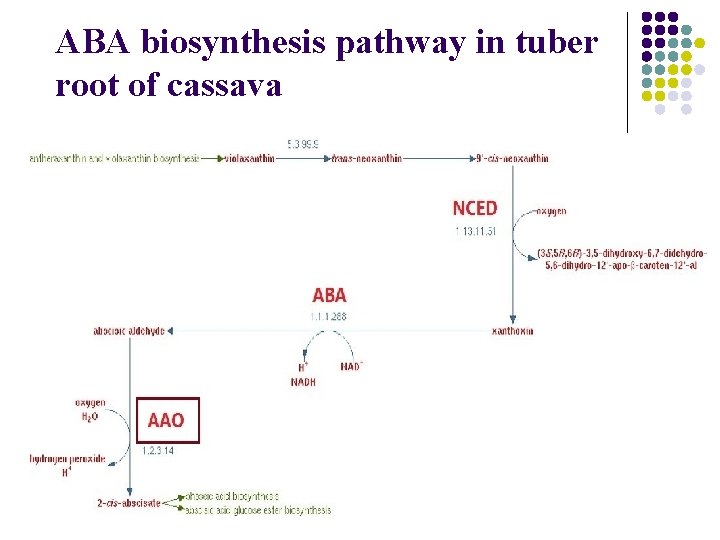
ABA biosynthesis pathway in tuber root of cassava

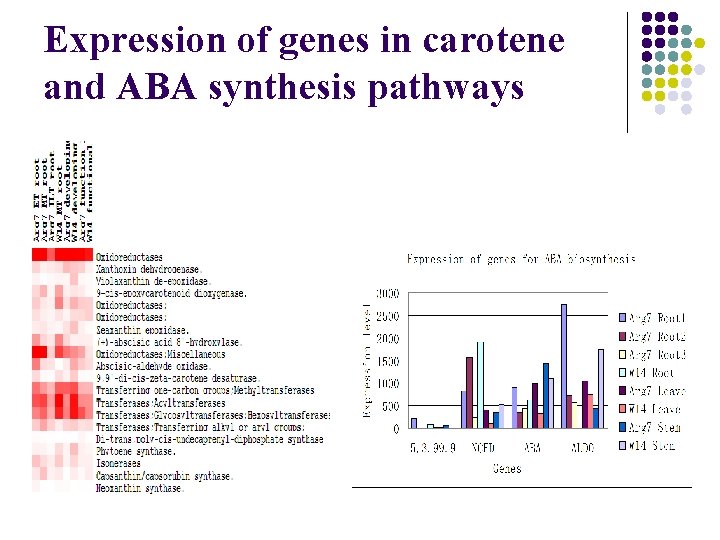
Expression of genes in carotene and ABA synthesis pathways

Comparative genomics among cassava, Jatropha and castor bean Unique gene families: Cassava 2043 Jatropha 532 Castor bean 826 Shared gene families: 12041

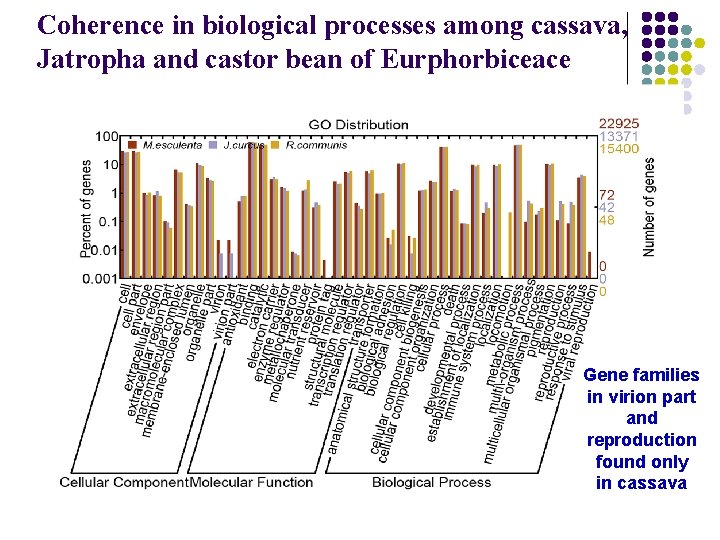
Coherence in biological processes among cassava, Jatropha and castor bean of Eurphorbiceace Gene families in virion part and reproduction found only in cassava

Database: Cassava-genome. cn

Ongoing work l l Genome fine mapping integrated assembly with physic map, BAC-end sequences and BAC-pooling sequences. Chromosomes location with assembling BACs and scarfolds based on in situ hybridization Functional verification of genes for important pathways Development of SNP markers and molecular design breeding

Summary l l l Genome drafts of an ancestor and a cultivar in cassava were assembled annotated. Genome diversity decreased from wild ancestor to cultivar in domestication; Millions of SNPs were discovered and recommended for genotyping in cassava. Advanced an efficient starch biosynthesis pathway in tuber root of cassava.

Acknowledgement CATAS BX Feng, Z Xia, XC Zhou, KM Li, PH Li, M Peng, WQ Wang l XJIEG-CAS Bin Liu, Binxiao Feng l SIS-CAS Jun Yang, Peng Zhang l BIG-CAS JF Xiao, JX Liu, SN Hu l Fudan U Zhicheng Wu, Ruiqi Liao, Shuigen Zhou l SCBG-CAS Gong Xiao, Chi Song, Ying Wang l l EMBRAPA, Brazil, Luiz C l UC Davis Mingcheng Luo Copenhagen U, Demark Rubini, Birger Muller l Nanjing Agricultural U Qunfeng Lu l
 Genome-to-genome distance calculator
Genome-to-genome distance calculator Cassava
Cassava Cassava project
Cassava project Cassava japanese
Cassava japanese Cassava
Cassava Sae criteria gcp
Sae criteria gcp Investigator responsibilities gcp
Investigator responsibilities gcp Gcp milan
Gcp milan Fda data integrity presentation
Fda data integrity presentation Documentation objectives
Documentation objectives Gcp project
Gcp project Gcp quality assurance
Gcp quality assurance The tokyo are on
The tokyo are on Gcp aiesec
Gcp aiesec Gcp regler
Gcp regler Sae gcp
Sae gcp The global health network gcp
The global health network gcp Global health network gcp
Global health network gcp Nihr gcp training
Nihr gcp training Protocol deviations gcp
Protocol deviations gcp Stanford genome technology center
Stanford genome technology center 1000 genome project
1000 genome project Chapter 14 the human genome
Chapter 14 the human genome Sequence assembly ppt
Sequence assembly ppt What is a human genome
What is a human genome National human genome research institute
National human genome research institute Igv genome browser
Igv genome browser Human genome features
Human genome features Human genome structure
Human genome structure What is genome
What is genome Genome.gov
Genome.gov Functional dna
Functional dna Genome klick
Genome klick Chapter 13 section 3 the human genome
Chapter 13 section 3 the human genome Human genome project source code
Human genome project source code Scale annotation pipeline
Scale annotation pipeline Human genome size
Human genome size Genome research limited
Genome research limited National human genome research institute
National human genome research institute