Functional Organization of the Transcriptome in Human Brain
Functional Organization of the Transcriptome in Human Brain Michael C. Oldham Laboratory of Daniel H. Geschwind, UCLA BIOCOMP ‘ 08, Las Vegas, NV July 15, 2008
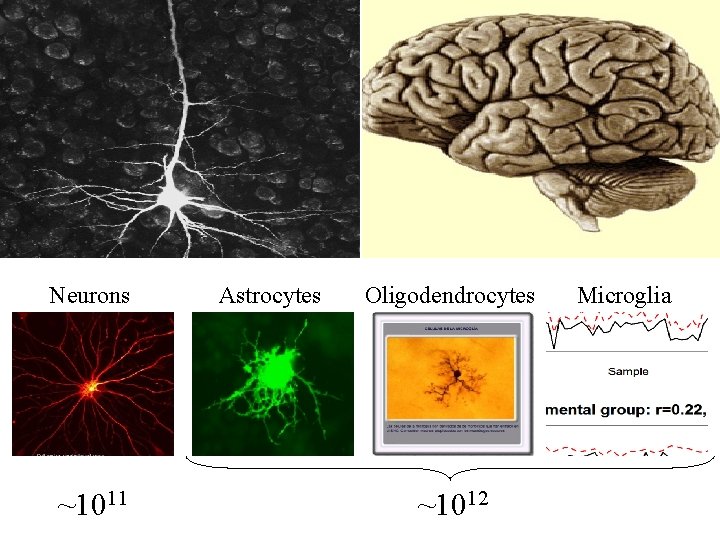
Neurons ~1011 Astrocytes Oligodendrocytes ~1012 Microglia
Microarray studies of the human brain face a number of challenges • Cellular heterogeneity – m. RNA extracted from tissue homogenates • Sample quality – Post-mortem tissue • Sample size – Typically, <10 individuals per group • Focus on differential expression
What’s wrong with differential expression?
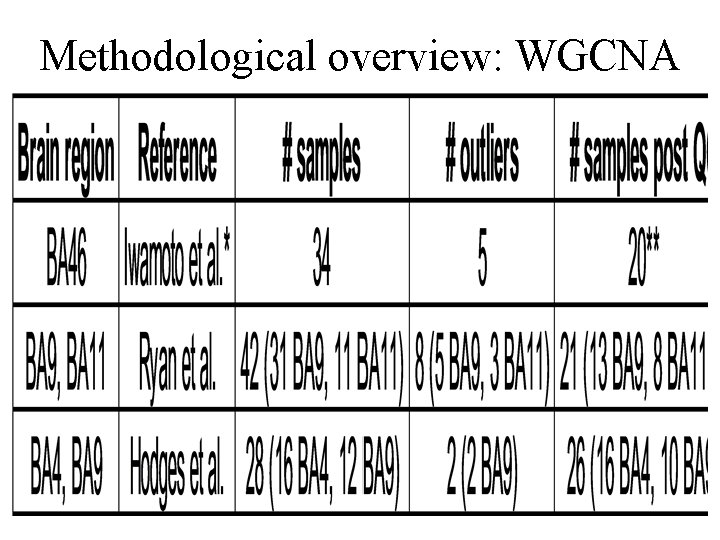
Methodological overview: WGCNA
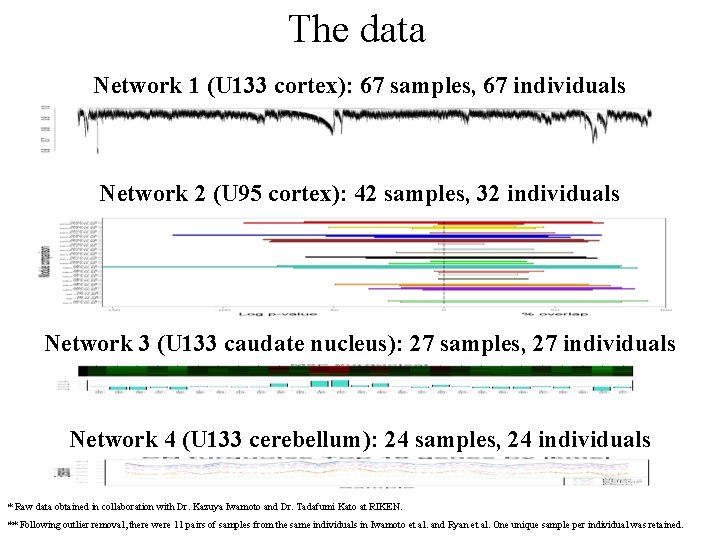
The data Network 1 (U 133 cortex): 67 samples, 67 individuals Network 2 (U 95 cortex): 42 samples, 32 individuals Network 3 (U 133 caudate nucleus): 27 samples, 27 individuals Network 4 (U 133 cerebellum): 24 samples, 24 individuals * Raw data obtained in collaboration with Dr. Kazuya Iwamoto and Dr. Tadafumi Kato at RIKEN. ** Following outlier removal, there were 11 pairs of samples from the same individuals in Iwamoto et al. and Ryan et al. One unique sample per individual was retained.
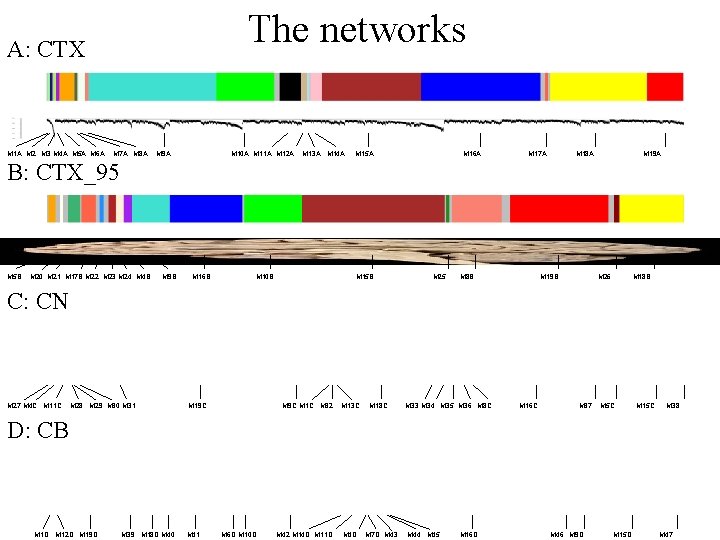
The networks A: CTX M 1 A M 2 M 3 M 4 A M 5 A M 6 A M 7 A M 8 A M 9 A M 10 A M 11 A M 12 A M 13 A M 14 A M 15 A M 16 A M 17 A M 18 A M 19 A B: CTX_95 M 5 B M 20 M 21 M 17 B M 22 M 23 M 24 M 4 B M 9 B M 16 B M 10 B M 15 B M 25 M 8 B M 19 B M 26 M 18 B C: CN M 27 M 4 C M 11 C M 28 M 29 M 30 M 31 M 19 C M 1 C M 32 M 13 C M 18 C M 33 M 34 M 35 M 36 M 8 C M 16 C M 37 M 5 C M 15 C M 38 D: CB M 1 D M 12 D M 19 D M 39 M 18 D M 40 M 41 M 6 D M 10 D M 42 M 14 D M 11 D M 4 D M 7 D M 43 M 44 M 45 M 16 D M 46 M 9 D M 15 D M 47
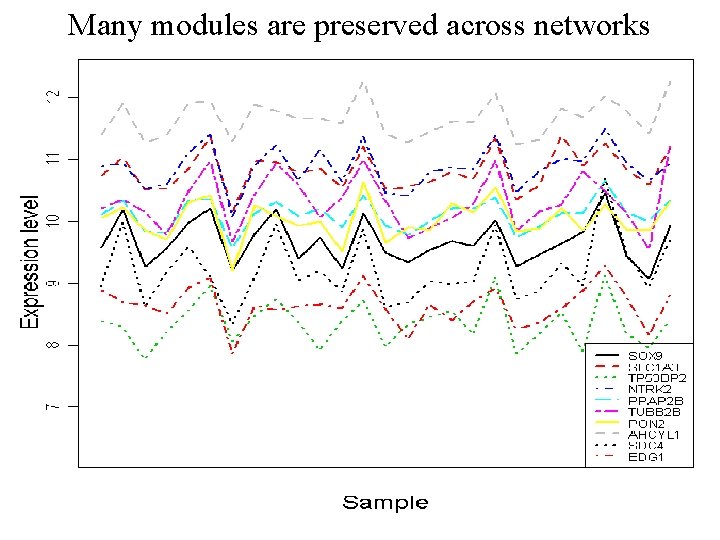
Many modules are preserved across networks
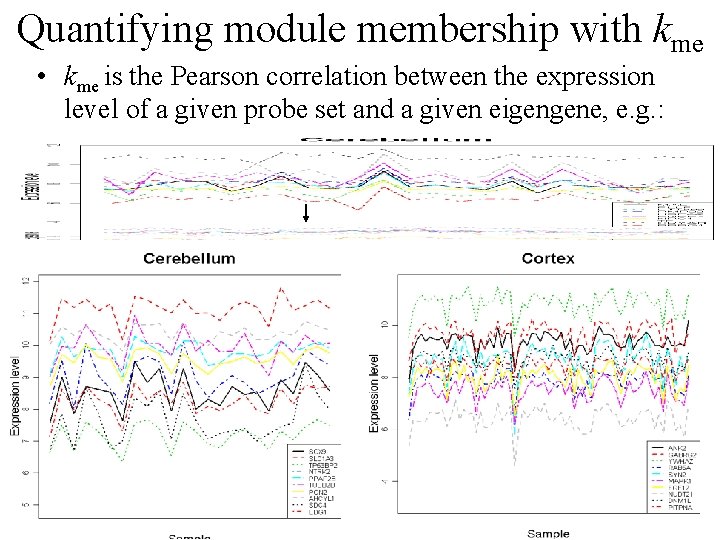
Quantifying module membership with kme • kme is the Pearson correlation between the expression level of a given probe set and a given eigengene, e. g. :

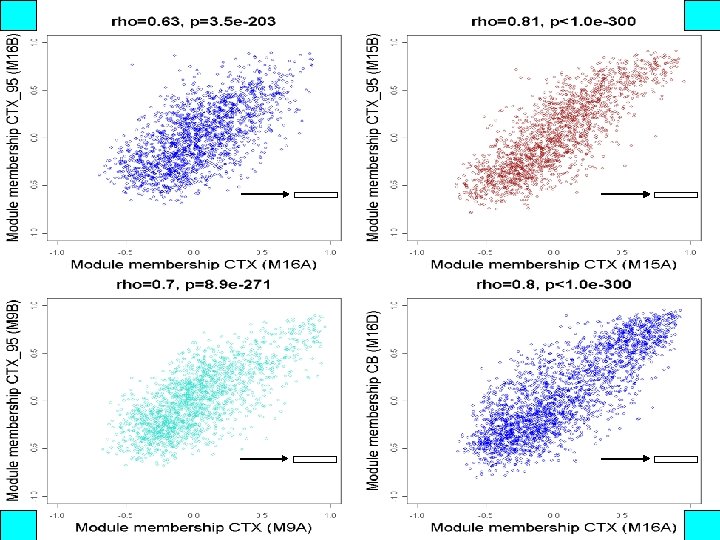
kme is significantly correlated across networks A B C
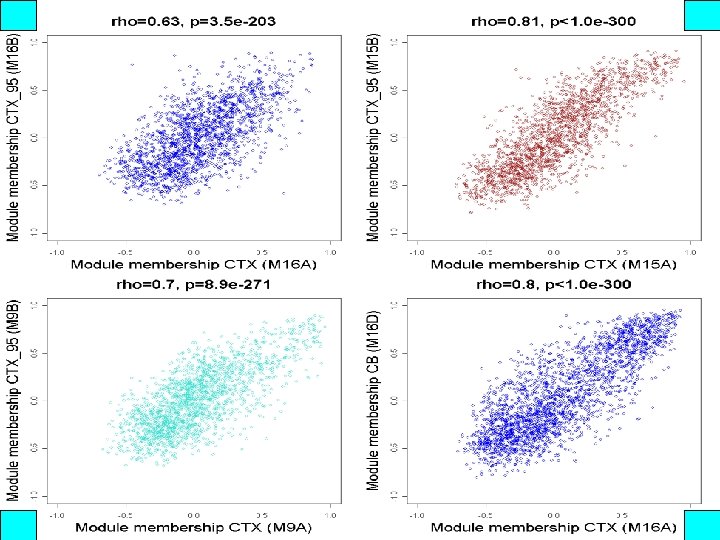
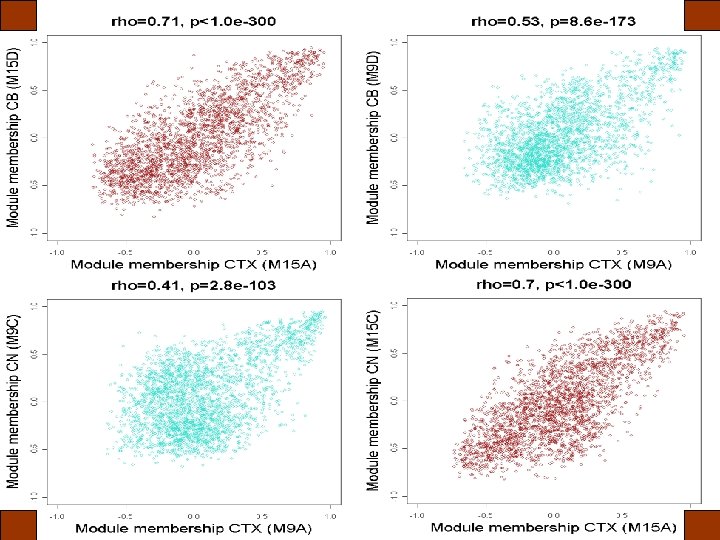
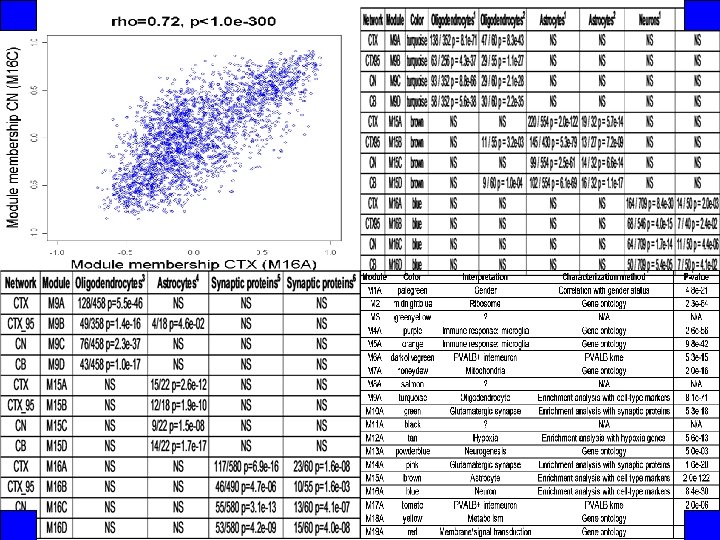
Conserved modules are enriched for markers of major cell classes 1 Cahoy, J. D. et al. J Neurosci 28: 264 -78 (2008) 3 Nielsen, J. A. et al. J Neurosci 26: 9881 -9891 (2006) 2 Lein, E. S. et al. Nature 445: 168 -76 (2007) 4 Bachoo, R. M. et al. PNAS 101: 8384 -8389 (2004) 5 Genes 2 Cognition Consortium 6 Morciano, M. et al. J Neurochem 95: 1732 -1745 (2005)
Amount Thought experiment Sample
The cortical transcriptome is organized into functional modules M 1 A M 2 M 3 M 4 A M 5 A M 6 A M 7 A M 8 A M 9 A M 10 A M 11 A M 12 A M 13 A M 14 A M 15 A M 16 A M 17 A M 18 A M 19 A
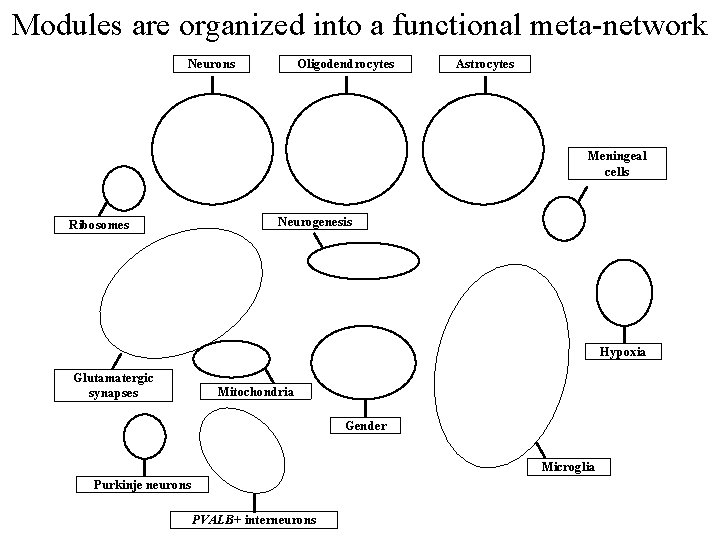
Modules are organized into a functional meta-network Neurons Oligodendrocytes Astrocytes Meningeal cells Ribosomes Neurogenesis Hypoxia Glutamatergic synapses Mitochondria Gender Microglia Purkinje neurons PVALB+ interneurons

Applications 1) Context-specific annotation for genes expressed in the human brain (“guilt-by-association”) • Rationale: genes with the strongest evidence of membership for the same module are likely to be driven by the same underlying factors
M 6 A: PVALB+ interneurons
Applications 1) Context-specific annotation for genes expressed in the human brain (“guilt-by-association”) 2) In silico comparisons of cellular specificity of gene expression across brain regions • Rationale: genes with the most significant differences in membership for cell-type modules between brain regions imply differences in the cellular specificity and/or consistency of gene expression
Applications 1) Context-specific annotation for genes expressed in the human brain (“guilt-by-association”) 2) In silico comparisons of cellular specificity of gene expression across brain regions 3) Cellular phenotype discovery • Rationale: unsupervised analysis of gene coexpression patterns can identify novel distinctions among cell types within brain regions
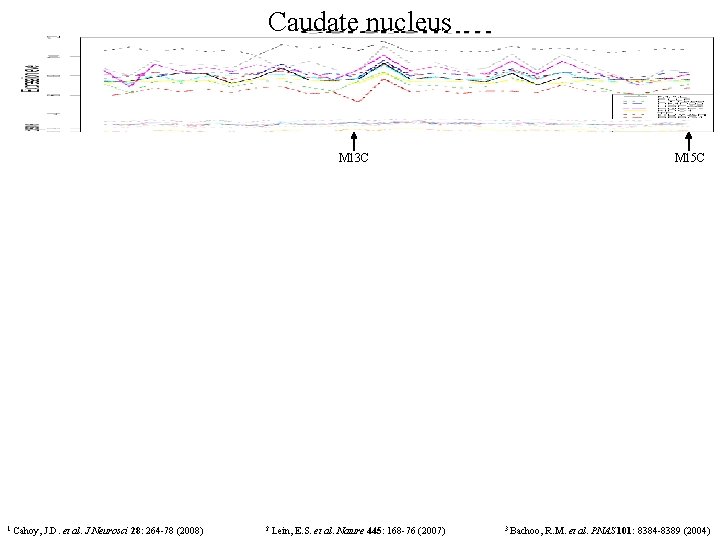
Caudate nucleus M 13 C 1 Cahoy, J. D. et al. J Neurosci 28: 264 -78 (2008) 2 Lein, E. S. et al. Nature 445: 168 -76 (2007) M 15 C 3 Bachoo, R. M. et al. PNAS 101: 8384 -8389 (2004)

M 13 C (CN) identifies genes that are coexpressed in the SVZ
Conclusions • The human brain transcriptome is organized into modules of coexpressed genes – • Several highly conserved modules are enriched for markers of major cell classes – – • Many modules are reproducible across microarrays, individuals, and brain regions ‘Core’ transcriptional programs for neurons, oligodendrocytes, astrocytes, and microglia Context-specific annotation for thousands of genes expressed in the human brain Potential to leverage consistency of kme for comparisons with other conditions of interest (e. g. disease)

Acknowledgements Dan Geschwind Steve Horvath Gena Konopka Peter Langfelder Collaborators: Dr. Kazuya Iwamoto Dr. Tadafumi Kato The Geschwind lab
- Slides: 28