Functional module identification with tomato gene and metabolite
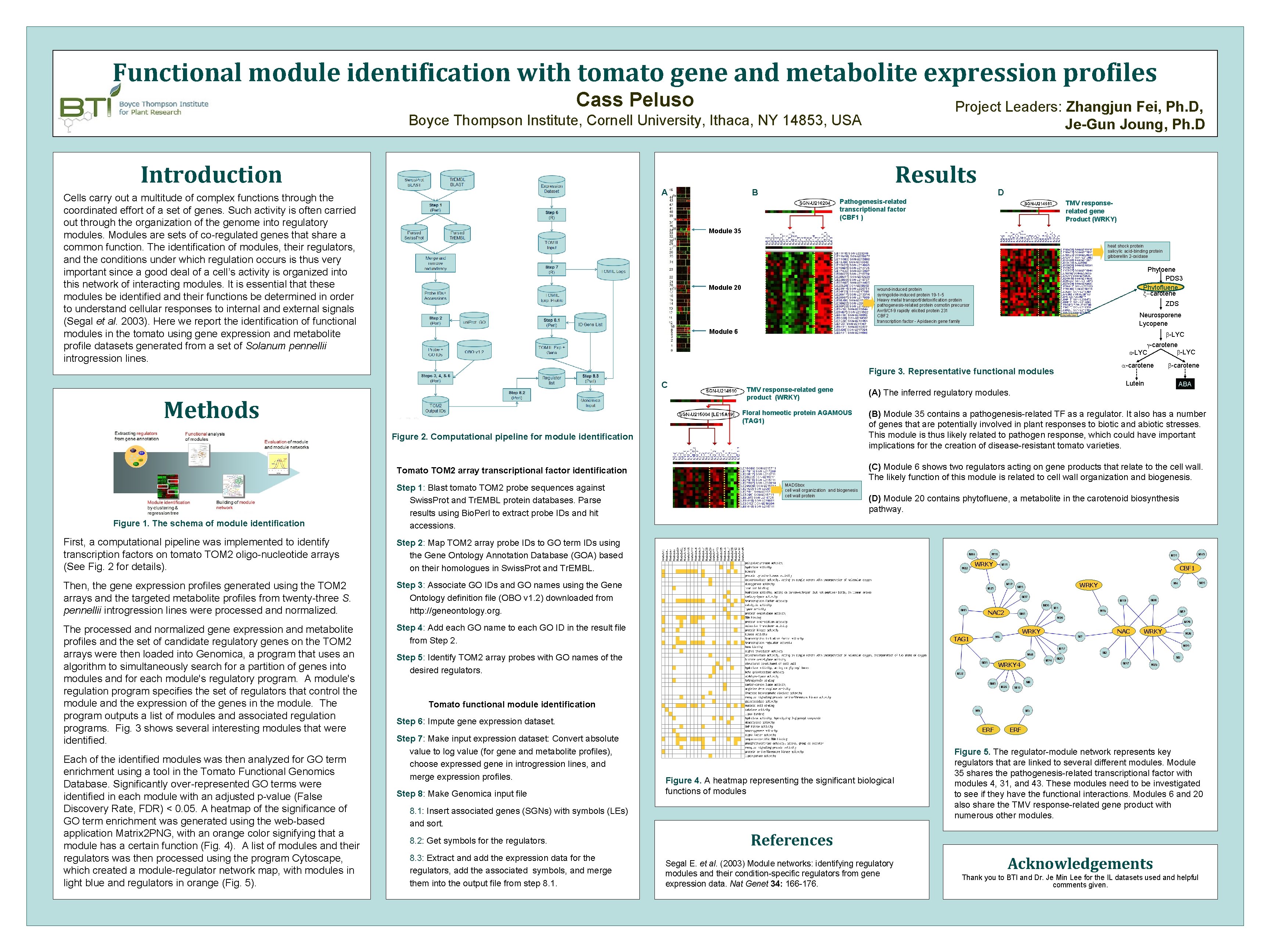
Functional module identification with tomato gene and metabolite expression profiles Cass Peluso Project Leaders: Zhangjun Fei, Ph. D, Je-Gun Joung, Ph. D Boyce Thompson Institute, Cornell University, Ithaca, NY 14853, USA Introduction Abstract Cells carry out a multitude of complex functions through the A Results B D Pathogenesis-related transcriptional factor (CBF 1 ) coordinated effort of a set of genes. Such activity is often carried out through the organization of the genome into regulatory modules. Modules are sets of co-regulated genes that share a common function. The identification of modules, their regulators, and the conditions under which regulation occurs is thus very important since a good deal of a cell’s activity is organized into this network of interacting modules. It is essential that these modules be identified and their functions be determined in order to understand cellular responses to internal and external signals (Segal et al. 2003). Here we report the identification of functional modules in the tomato using gene expression and metabolite profile datasets generated from a set of Solanum pennellii introgression lines. TMV responserelated gene Product (WRKY) Module 35 heat shock protein salicylic acid-binding protein gibberellin 2 -oxidase Module 20 Phytoene PDS 3 Phytofluene x-carotene wound-induced protein syringolide-induced protein 19 -1 -5 Heavy metal transport/detoxification protein pathogenesis-related protein osmotin precursor Avr 9/Cf-9 rapidly elicited protein 231 CBF 2 transcription factor - Apidaecin gene family ZDS Neurosporene Lycopene Module 6 b-LYC g-carotene b-LYC e-LYC a-carotene Figure 3. Representative functional modules C Methods TMV response-related gene product (WRKY) Floral homeotic protein AGAMOUS (TAG 1) Figure 2. Computational pipeline for module identification Figure 1. The schema of module identification First, a computational pipeline was implemented to identify transcription factors on tomato TOM 2 oligo-nucleotide arrays (See Fig. 2 for details). Step 2: Map TOM 2 array probe IDs to GO term IDs using the Gene Ontology Annotation Database (GOA) based on their homologues in Swiss. Prot and Tr. EMBL. Then, the gene expression profiles generated using the TOM 2 arrays and the targeted metabolite profiles from twenty-three S. pennellii introgression lines were processed and normalized. Step 3: Associate GO IDs and GO names using the Gene Ontology definition file (OBO v 1. 2) downloaded from http: //geneontology. org. The processed and normalized gene expression and metabolite profiles and the set of candidate regulatory genes on the TOM 2 arrays were then loaded into Genomica, a program that uses an algorithm to simultaneously search for a partition of genes into modules and for each module's regulatory program. A module's regulation program specifies the set of regulators that control the module and the expression of the genes in the module. The program outputs a list of modules and associated regulation programs. Fig. 3 shows several interesting modules that were identified. Step 4: Add each GO name to each GO ID in the result file from Step 2. Each of the identified modules was then analyzed for GO term enrichment using a tool in the Tomato Functional Genomics Database. Significantly over-represented GO terms were identified in each module with an adjusted p-value (False Discovery Rate, FDR) < 0. 05. A heatmap of the significance of GO term enrichment was generated using the web-based application Matrix 2 PNG, with an orange color signifying that a module has a certain function (Fig. 4). A list of modules and their regulators was then processed using the program Cytoscape, which created a module-regulator network map, with modules in light blue and regulators in orange (Fig. 5). (A) The inferred regulatory modules. ABA (B) Module 35 contains a pathogenesis-related TF as a regulator. It also has a number of genes that are potentially involved in plant responses to biotic and abiotic stresses. This module is thus likely related to pathogen response, which could have important implications for the creation of disease-resistant tomato varieties. (C) Module 6 shows two regulators acting on gene products that relate to the cell wall. The likely function of this module is related to cell wall organization and biogenesis. Tomato TOM 2 array transcriptional factor identification Step 1: Blast tomato TOM 2 probe sequences against Swiss. Prot and Tr. EMBL protein databases. Parse results using Bio. Perl to extract probe IDs and hit accessions. Lutein b-carotene MADSbox cell wall organization and biogenesis cell wall protein (D) Module 20 contains phytofluene, a metabolite in the carotenoid biosynthesis pathway. WRKY CBF 1 WRKY NAC 2 WRKY NAC WRKY TAG 1 Step 5: Identify TOM 2 array probes with GO names of the desired regulators. WRKY 4 Tomato functional module identification Step 6: Impute gene expression dataset. Step 7: Make input expression dataset: Convert absolute value to log value (for gene and metabolite profiles), choose expressed gene in introgression lines, and merge expression profiles. Step 8: Make Genomica input file ERF Figure 4. A heatmap representing the significant biological functions of modules 8. 1: Insert associated genes (SGNs) with symbols (LEs) and sort. 8. 2: Get symbols for the regulators. 8. 3: Extract and add the expression data for the regulators, add the associated symbols, and merge them into the output file from step 8. 1. ERF Figure 5. The regulator-module network represents key regulators that are linked to several different modules. Module 35 shares the pathogenesis-related transcriptional factor with modules 4, 31, and 43. These modules need to be investigated to see if they have the functional interactions. Modules 6 and 20 also share the TMV response-related gene product with numerous other modules. References Segal E. et al. (2003) Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet 34: 166 -176. Acknowledgements Thank you to BTI and Dr. Je Min Lee for the IL datasets used and helpful comments given.
- Slides: 1