Functional Genomics with NextGeneration Sequencing Jen Taylor Bioinformatics

Functional Genomics with Next-Generation Sequencing Jen Taylor Bioinformatics Team CSIRO Plant Industry
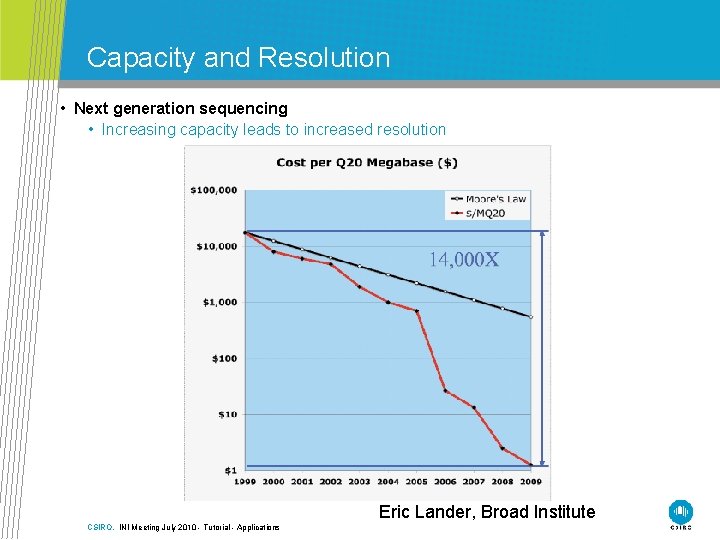
Capacity and Resolution • Next generation sequencing • Increasing capacity leads to increased resolution Eric Lander, Broad Institute CSIRO. INI Meeting July 2010 - Tutorial - Applications
How a Genome Works? Parts Description • Function? • Interconnectedness? Comparisons • Population - level • Between genomes CSIRO. INI Meeting July 2010 - Tutorial - Applications
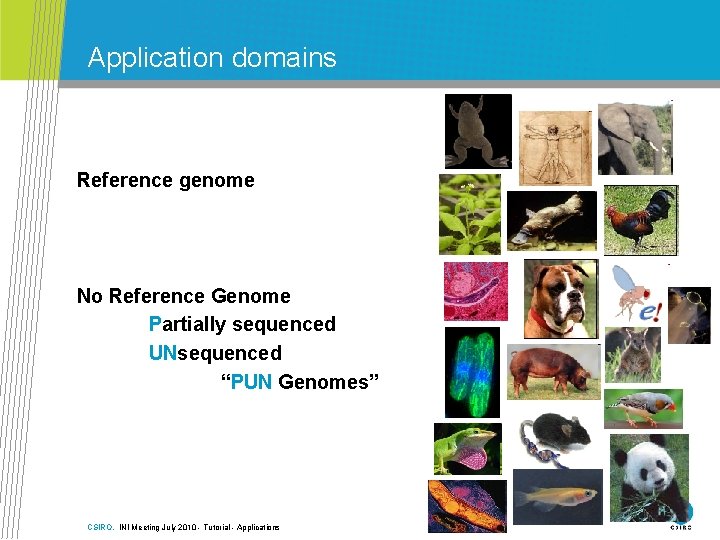
Application domains Reference genome No Reference Genome Partially sequenced UNsequenced “PUN Genomes” CSIRO. INI Meeting July 2010 - Tutorial - Applications

Impact of a Reference Genome Sequence Data Genome Alignment Assembly Read Density Contigs Characterisation CSIRO. INI Meeting July 2010 - Tutorial - Applications
Applications of Next Generation Sequencing • Profiling of Variation • • • Discovery Genetic variation Transcript variation Epigenetic variation Metagenomic variation • • Novel genomes Novel genes Novel transcripts Small / long non-coding RNA Today RNA Sequencing (RNASeq) • Coding and non-coding transcript profiling • Dynamic and Context dependent Epigenomics • Genome-wide protein-DNA interactions, DNA modifications • Heritable and reversible regulation of gene expression CSIRO. INI Meeting July 2010 - Tutorial - Applications
RNASeq • Qualitative – transcript diversity • Quantitative – transcript abundance • Impact of NGS • Observation of transcript complexity • Transcript discovery • Small / long non-coding RNA • Analytical challenges • Transcript complexity • Compositional properties CSIRO. INI Meeting July 2010 - Tutorial - Applications
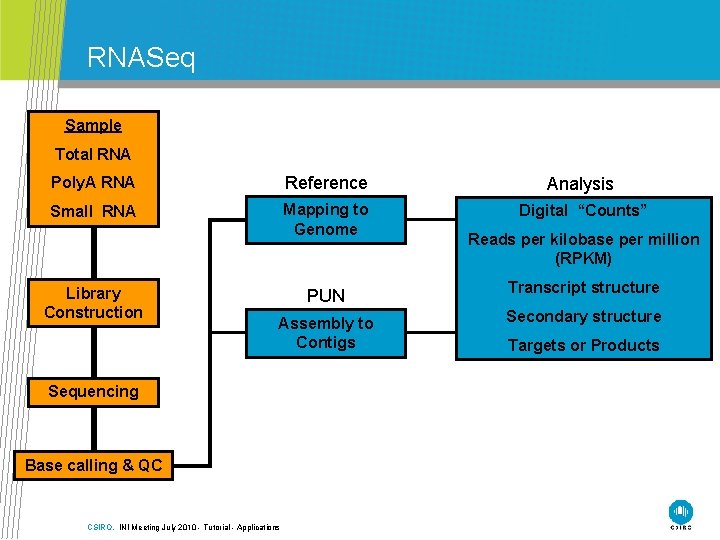
RNASeq Sample Total RNA Poly. A RNA Reference Analysis Small RNA Mapping to Genome Digital “Counts” Library Construction Reads per kilobase per million (RPKM) PUN Transcript structure Assembly to Contigs Secondary structure Sequencing Base calling & QC CSIRO. INI Meeting July 2010 - Tutorial - Applications Targets or Products
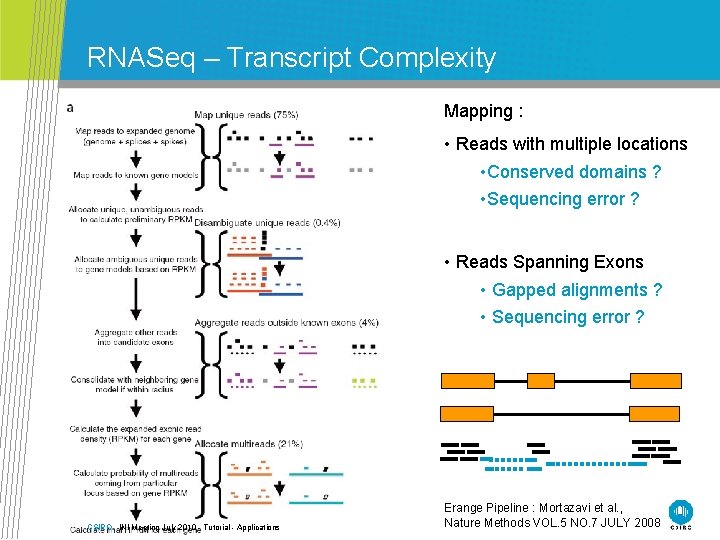
RNASeq – Transcript Complexity Mapping : • Reads with multiple locations • Conserved domains ? • Sequencing error ? • Reads Spanning Exons • Gapped alignments ? • Sequencing error ? CSIRO. INI Meeting July 2010 - Tutorial - Applications Erange Pipeline : Mortazavi et al. , Nature Methods VOL. 5 NO. 7 JULY 2008
RNASeq – Compositional properties Depth of Sequence • Sequence count ≈ Transcript Abundance • Majority of the data can be dominated by a small number of highly abundant transcripts • Ability to observe transcripts of smaller abundance is dependent upon sequence depth CSIRO. INI Meeting July 2010 - Tutorial - Applications
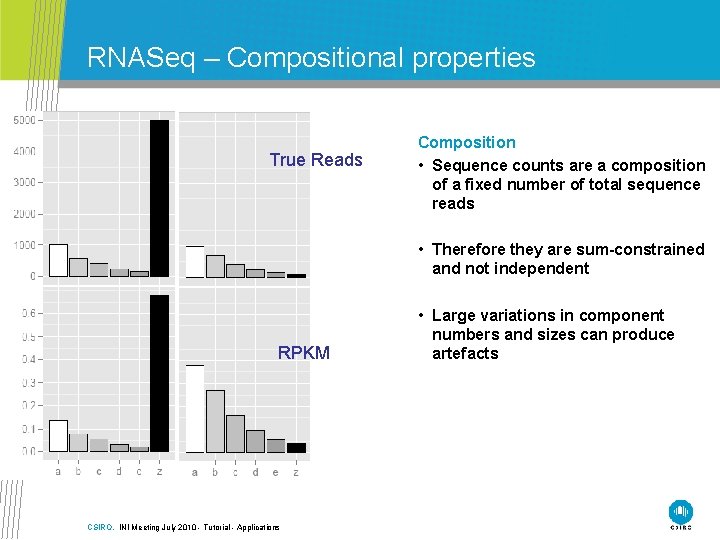
RNASeq – Compositional properties True Reads Composition • Sequence counts are a composition of a fixed number of total sequence reads • Therefore they are sum-constrained and not independent RPKM CSIRO. INI Meeting July 2010 - Tutorial - Applications • Large variations in component numbers and sizes can produce artefacts
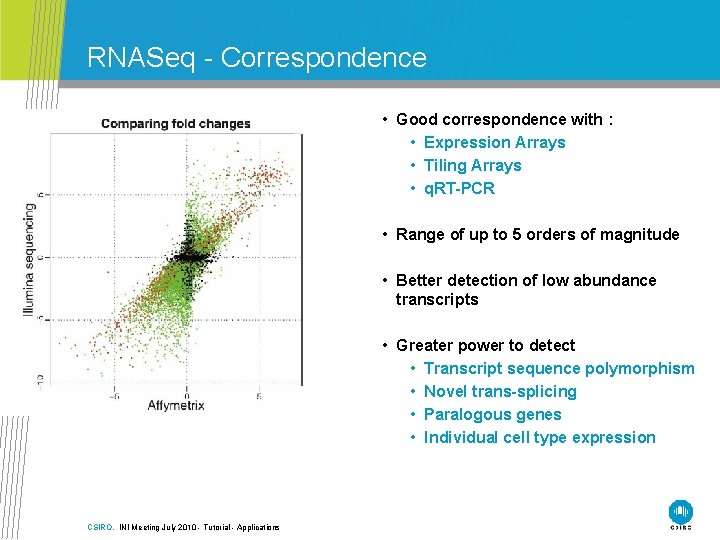
RNASeq - Correspondence • Good correspondence with : • Expression Arrays • Tiling Arrays • q. RT-PCR • Range of up to 5 orders of magnitude • Better detection of low abundance transcripts • Greater power to detect • Transcript sequence polymorphism • Novel trans-splicing • Paralogous genes • Individual cell type expression CSIRO. INI Meeting July 2010 - Tutorial - Applications
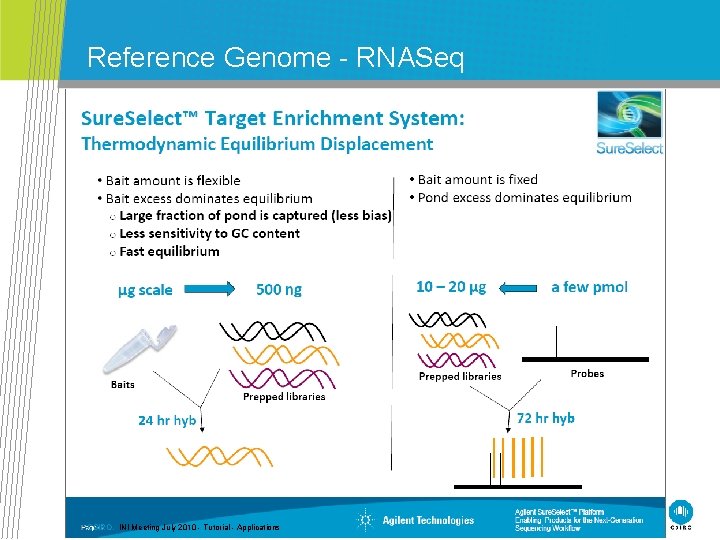
Reference Genome - RNASeq CSIRO. INI Meeting July 2010 - Tutorial - Applications
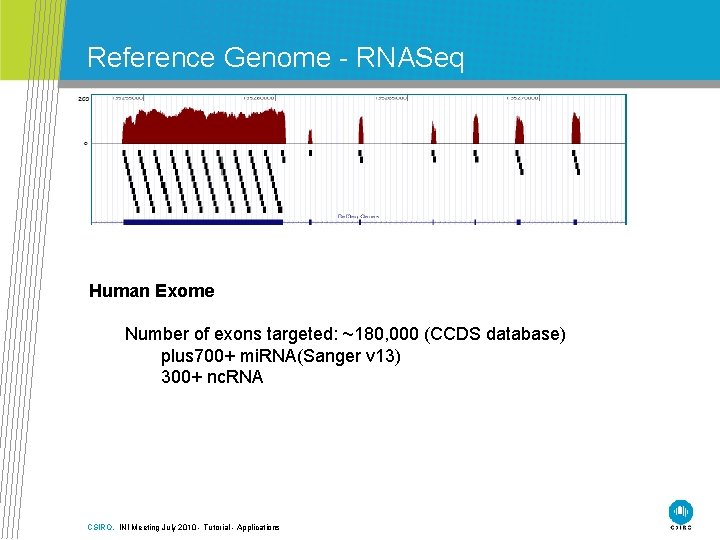
Reference Genome - RNASeq Human Exome Number of exons targeted: ~180, 000 (CCDS database) plus 700+ mi. RNA(Sanger v 13) 300+ nc. RNA CSIRO. INI Meeting July 2010 - Tutorial - Applications
![Epigenome • Protein-DNA interactions [Ch. IPSeq] • Nucleosome positioning • Histone modification • Transcription Epigenome • Protein-DNA interactions [Ch. IPSeq] • Nucleosome positioning • Histone modification • Transcription](http://slidetodoc.com/presentation_image_h2/a807e84b189cb21b430d39f3caf13b92/image-15.jpg)
Epigenome • Protein-DNA interactions [Ch. IPSeq] • Nucleosome positioning • Histone modification • Transcription factor interactions • Methylation [Methyl. Seq] • Impact of Next. Gen • Whole genome profiling • Resolution • Analytical challenges • Systematic bias • Unambiguous mapping • Robust event calling CSIRO. INI Meeting July 2010 - Tutorial - Applications Image : Clear. Science
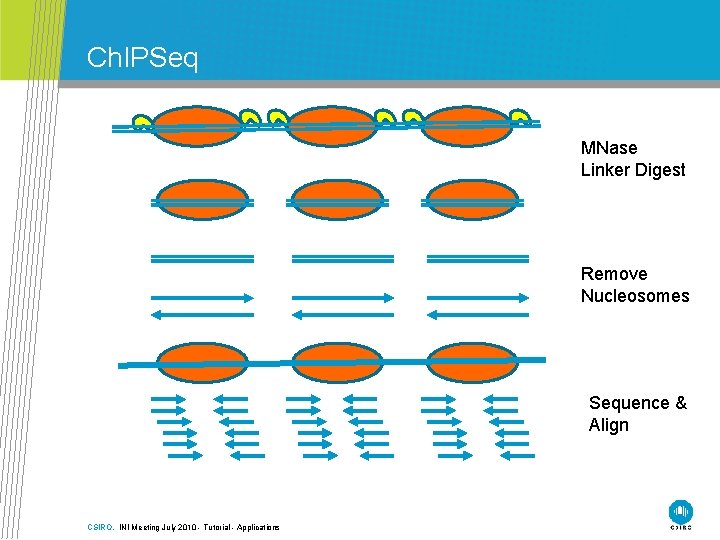
Ch. IPSeq MNase Linker Digest Remove Nucleosomes Sequence & Align CSIRO. INI Meeting July 2010 - Tutorial - Applications
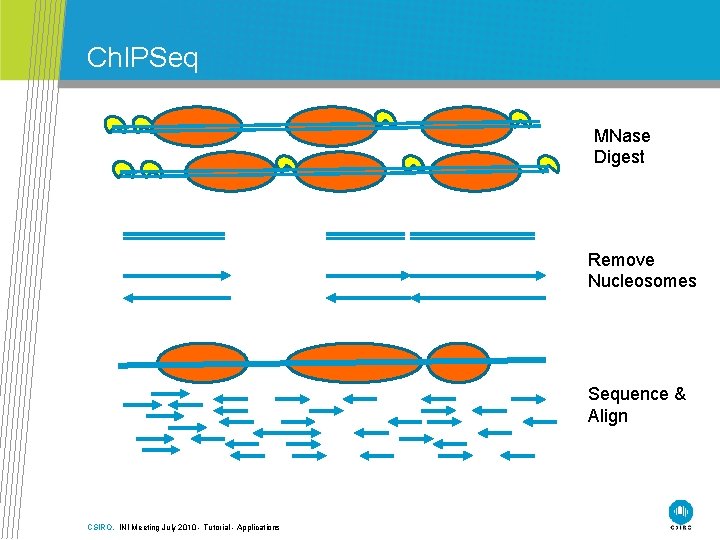
Ch. IPSeq MNase Digest Remove Nucleosomes Sequence & Align CSIRO. INI Meeting July 2010 - Tutorial - Applications
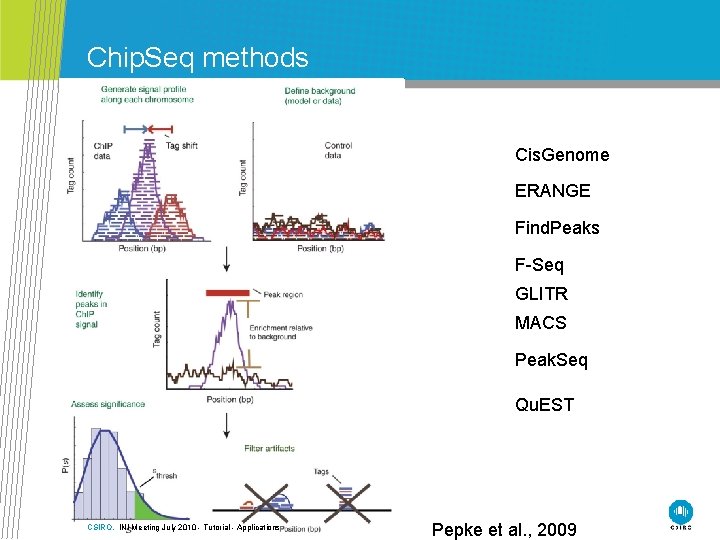
Chip. Seq methods Cis. Genome ERANGE Find. Peaks F-Seq GLITR MACS Peak. Seq Qu. EST CSIRO. INI Meeting July 2010 - Tutorial - Applications Pepke et al. , 2009
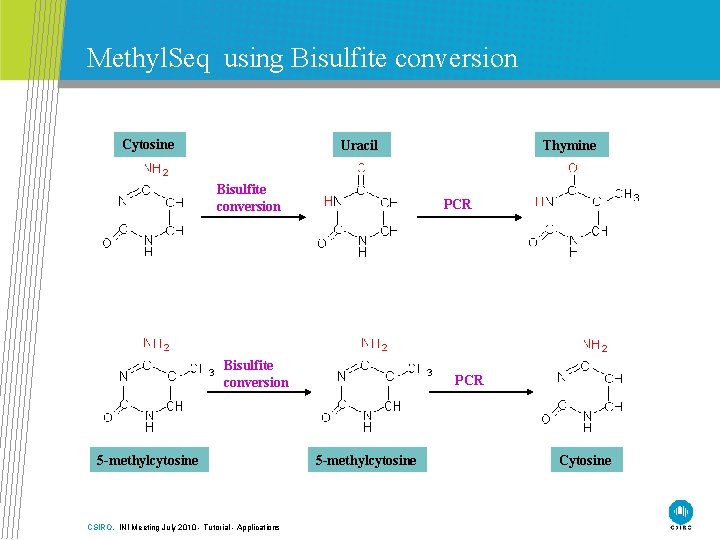
Methyl. Seq using Bisulfite conversion Cytosine Uracil Bisulfite conversion PCR Bisulfite conversion 5 -methylcytosine CSIRO. INI Meeting July 2010 - Tutorial - Applications Thymine PCR 5 -methylcytosine Cytosine
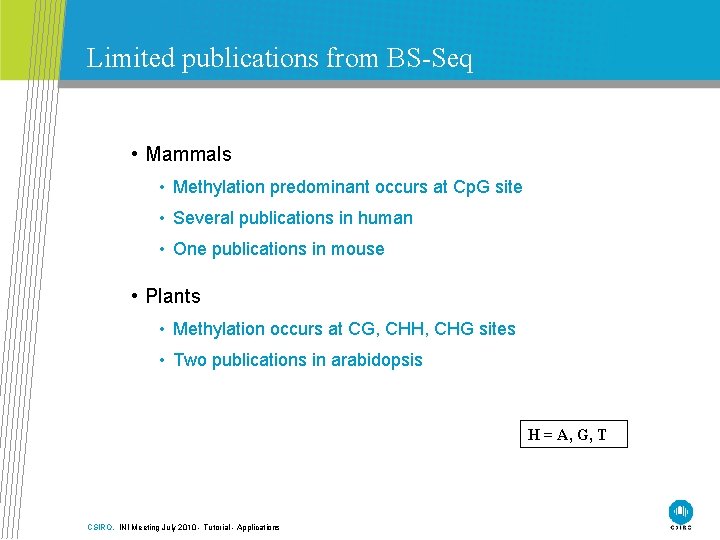
Limited publications from BS-Seq • Mammals • Methylation predominant occurs at Cp. G site • Several publications in human • One publications in mouse • Plants • Methylation occurs at CG, CHH, CHG sites • Two publications in arabidopsis H = A, G, T CSIRO. INI Meeting July 2010 - Tutorial - Applications
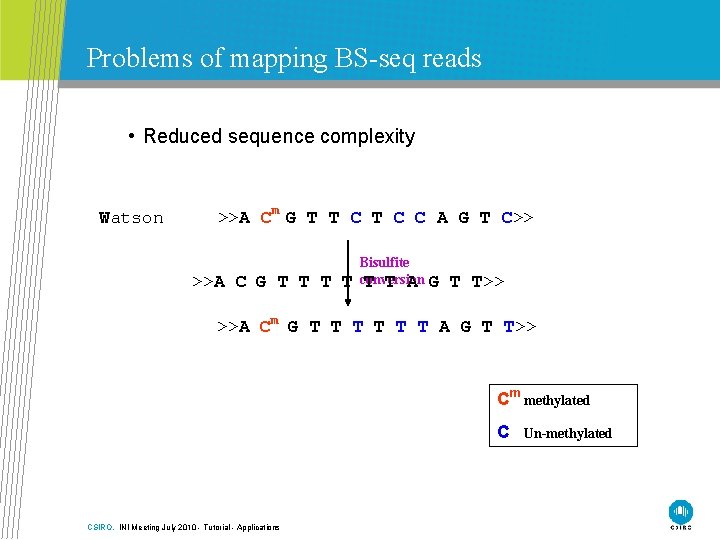
Problems of mapping BS-seq reads • Reduced sequence complexity Watson >>A Cm G T T C C A G T C>> >>A C G T T Bisulfite conversion T T A G T T>> >>A Cm G T T T A G T T>> Cm methylated C CSIRO. INI Meeting July 2010 - Tutorial - Applications Un-methylated
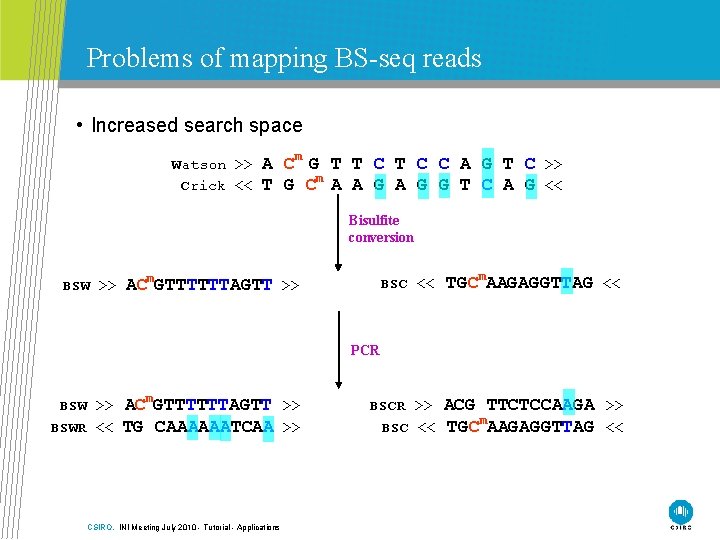
Problems of mapping BS-seq reads • Increased search space Watson >> Crick << A Cm G T T C C A G T C >> T G Cm A A G G T C A G << Bisulfite conversion BSW >> BSC << TGCm. AAGAGGTTAG << BSCR >> BSC << ACG TTCTCCAAGA >> TGCm. AAGAGGTTAG << ACm. GTTTTTTAGTT >> PCR BSW >> BSWR << ACm. GTTTTTTAGTT >> TG CAAAAAATCAA >> CSIRO. INI Meeting July 2010 - Tutorial - Applications

ELAND • Mapping reads to genome sequences • Mapping reads to two converted genome sequences • Cross match for reads mapping to multiple positions in converted genomes • Mapping results were combined to generate methylation information • Eland only allows 2 mismatches. Lister et al. Cell (2008) CSIRO. INI Meeting July 2010 - Tutorial - Applications
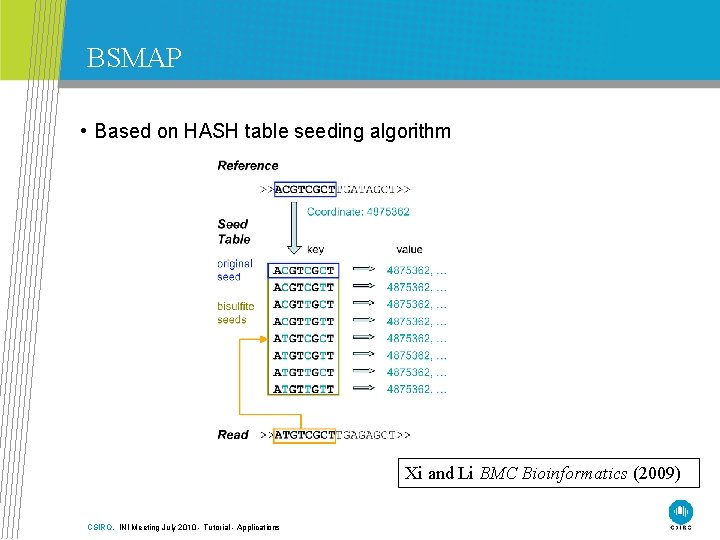
BSMAP • Based on HASH table seeding algorithm Xi and Li BMC Bioinformatics (2009) CSIRO. INI Meeting July 2010 - Tutorial - Applications
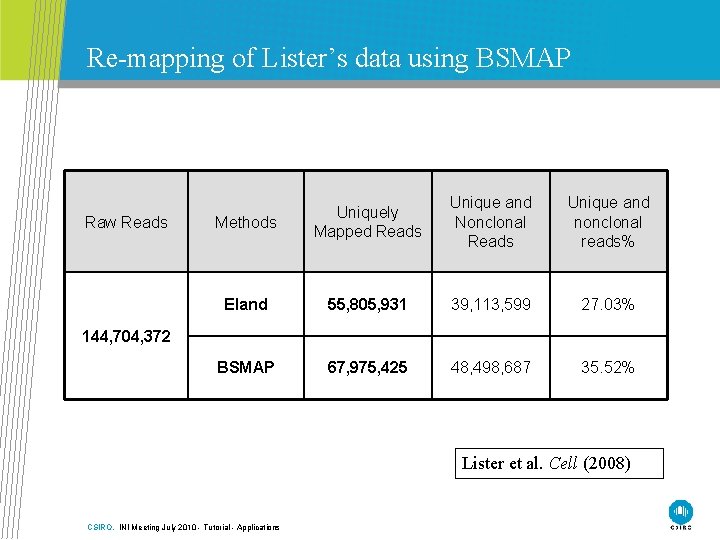
Re-mapping of Lister’s data using BSMAP Raw Reads Methods Uniquely Mapped Reads Unique and Nonclonal Reads Unique and nonclonal reads% Eland 55, 805, 931 39, 113, 599 27. 03% BSMAP 67, 975, 425 48, 498, 687 35. 52% 144, 704, 372 Lister et al. Cell (2008) CSIRO. INI Meeting July 2010 - Tutorial - Applications
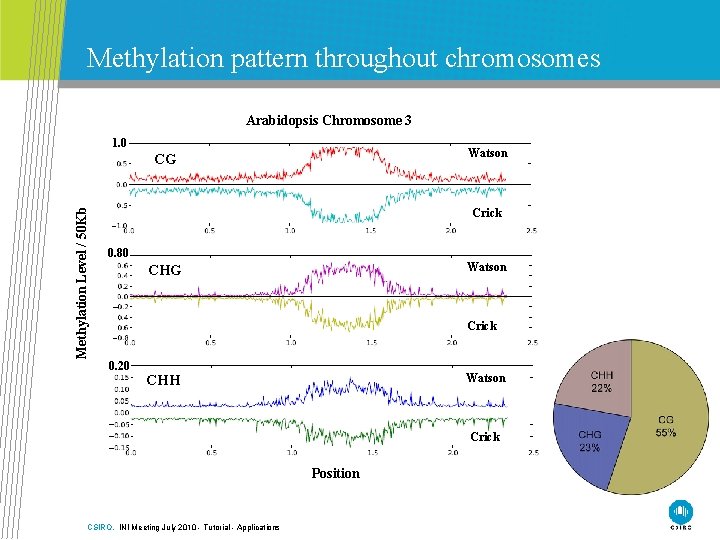
Methylation pattern throughout chromosomes Arabidopsis Chromosome 3 1. 0 Watson Methylation Level / 50 Kb CG Crick 0. 80 Watson CHG Crick 0. 20 CHH Watson Crick Position CSIRO. INI Meeting July 2010 - Tutorial - Applications
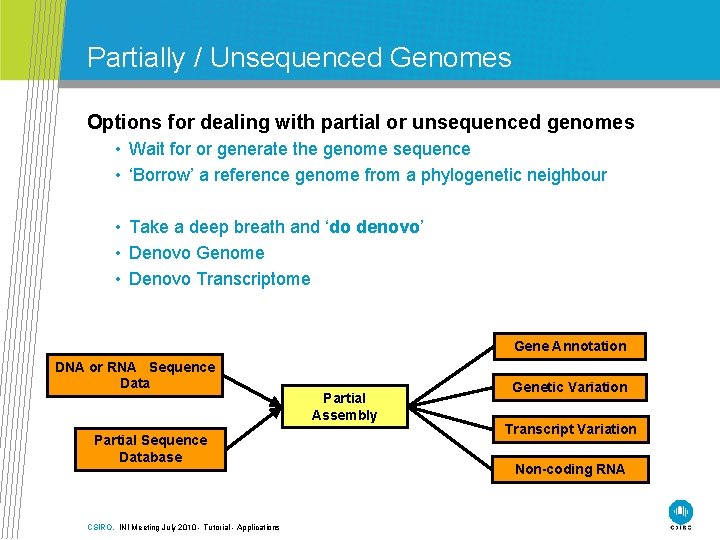
Partially / Unsequenced Genomes Options for dealing with partial or unsequenced genomes • Wait for or generate the genome sequence • ‘Borrow’ a reference genome from a phylogenetic neighbour • Take a deep breath and ‘do denovo’ • Denovo Genome • Denovo Transcriptome Gene Annotation DNA or RNA Sequence Data Partial Sequence Database CSIRO. INI Meeting July 2010 - Tutorial - Applications Partial Assembly Genetic Variation Transcript Variation Non-coding RNA
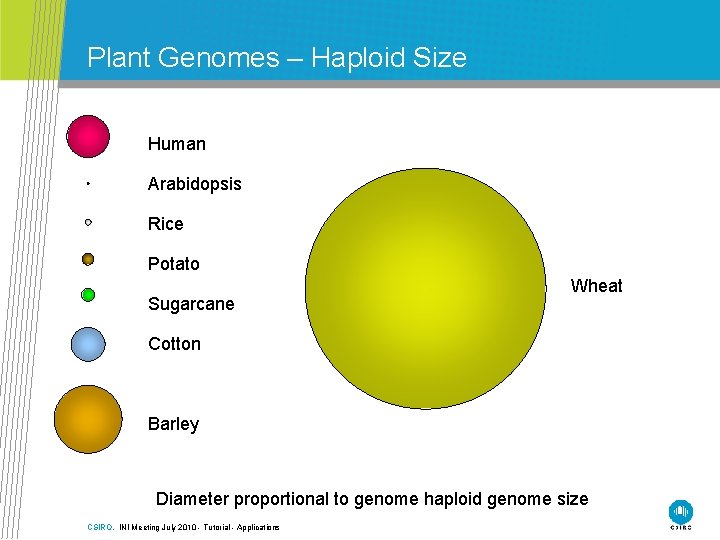
Plant Genomes – Haploid Size Human Arabidopsis Rice Potato Sugarcane Wheat Cotton Barley Diameter proportional to genome haploid genome size CSIRO. INI Meeting July 2010 - Tutorial - Applications
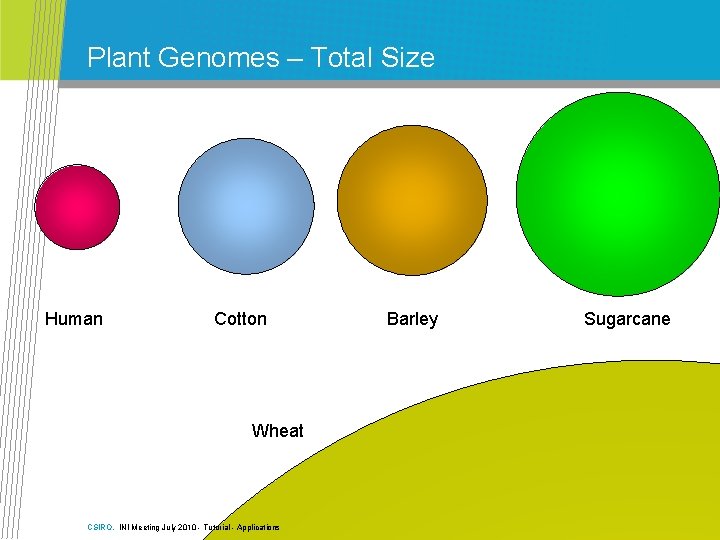
Plant Genomes – Total Size Human Cotton Wheat CSIRO. INI Meeting July 2010 - Tutorial - Applications Barley Sugarcane
Denovo RNA Seq • Why transcriptome ? • Large genome sizes with high repeat content are difficult to assemble • Transcriptomes more constant size • Enriched for functional content • Aims : • Transcript discovery • Small /long non-coding RNA profiling • Analytical challenges • Assembly – ABy. SS, Velvet, Euler-SR • Comparisons between non-discrete, overlapping transcripts • Annotation • Ploidy CSIRO. INI Meeting July 2010 - Tutorial - Applications
Summary – Impacts and Challenges • RNASeq • • Increased resolution Increased power for transcript complexity and variation Analytical challenges – transcript complexity, compositional bias Large gains in small and long non-coding RNA profiling • Epigenomics • Chip. Seq and Methyl. Seq • Genome-wide with resolution • Robust event calling is challenging • Denovo transcriptomics • Attractive option for large, repeat rich genomes CSIRO. INI Meeting July 2010 - Tutorial - Applications

Acknowledgements CSIRO PI Bioinformatics Team Andrew Spriggs Stuart Stephen Emily Ying Jose Robles Michael James CSIRO Biostatistics David Lovell CSIRO. INI Meeting July 2010 - Tutorial - Applications
- Slides: 32