FUNCTIONAL ANALYSIS OF PROTEIN SEQUENCES ANNOTATION AND FAMILY











- Slides: 54

FUNCTIONAL ANALYSIS OF PROTEIN SEQUENCES: ANNOTATION AND FAMILY CLASSIFICATION Anastasia Nikolskaya PIR (Protein Information Resource), Georgetown University Medical Center

Problem: l l l Overview Most new protein sequences come from genome sequencing projects Many have unknown functions Large-scale functional annotation of these sequences based simply on BLAST best hit has pitfalls; results are far from perfect Functional Analysis of Protein Sequences: l l Homology-based (sequence analysis, structure analysis) Non-homology (genome context, phylogenetic distribution) Solution for Large-scale Annotation: l l Highly curated annotated protein classification system Automatic annotation of sequences based on protein families PIRSF Protein Classification System l l Whole-protein family classification based on evolution Highly annotated, optimized for annotation propagation Functional predictions for uncharacterized proteins Used to facilitate and standardize annotations in Uni. Prot 2

Proteomics and Bioinformatics l Data: Gene expression profiling Genome-wide analysis of gene expression l l Data: Protein-protein interaction Data: Structural genomics 3 D structures of all protein families l l Data: Genome projects (Sequencing) …. Bioinformatics Computational analysis and integration of these data Making predictions (function etc), reconstructing 3 pathways

What’s In It For Me? l l When an experiment yields a sequence (or a set of sequences), we need to find out as much as we can about this protein and its possible function from available data Especially important for poorly characterized or uncharacterized (“hypothetical”) proteins More challenging for large sets of sequences generated by large-scale proteomics experiments The quality of this assessment is often critical for interpreting experimental results and making hypothesis for future experiments Sequence function 4

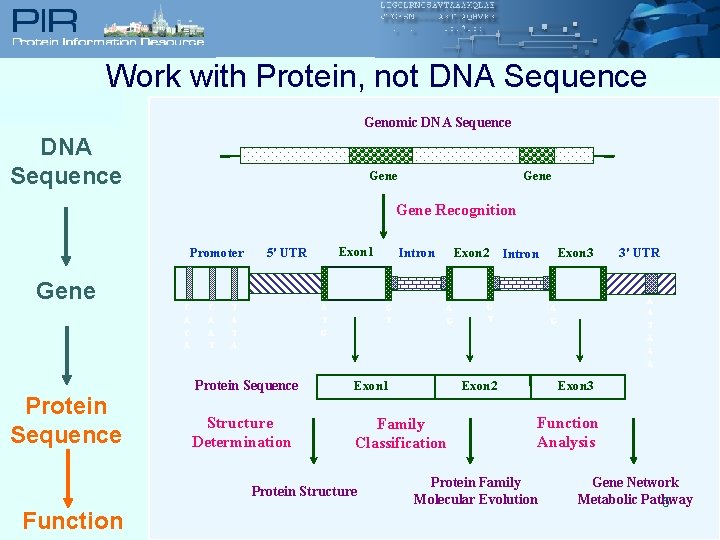
Work with Protein, not DNA Sequence Genomic DNA Sequence Gene Recognition Promoter Gene Protein Sequence C A C A A T Exon 1 5' UTR T A A T G T Exon 2 A G Exon 3 Intron G T Protein Sequence Exon 1 Structure Determination Family Classification Exon 2 3' UTR A A T A A G G Protein Structure Function Intron Exon 3 Function Analysis Protein Family Molecular Evolution Gene Network Metabolic Pathway 5

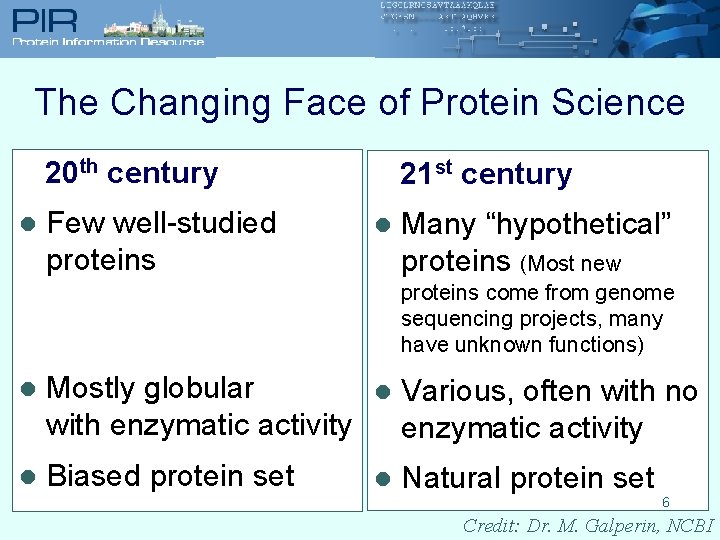
The Changing Face of Protein Science 20 th century l Few well-studied proteins 21 st century l Many “hypothetical” proteins (Most new proteins come from genome sequencing projects, many have unknown functions) l Mostly globular with enzymatic activity l Various, often with no enzymatic activity l Biased protein set l Natural protein set 6 Credit: Dr. M. Galperin, NCBI

Knowing the Complete Genome Sequence Advantages: l l All encoded proteins can be predicted and identified The missing functions can be identified analyzed Peculiarities and novelties in each organism can be studied Predictions can be made and verified Challenge: l Accurate assignment of known or predicted functions (functional annotation) 7

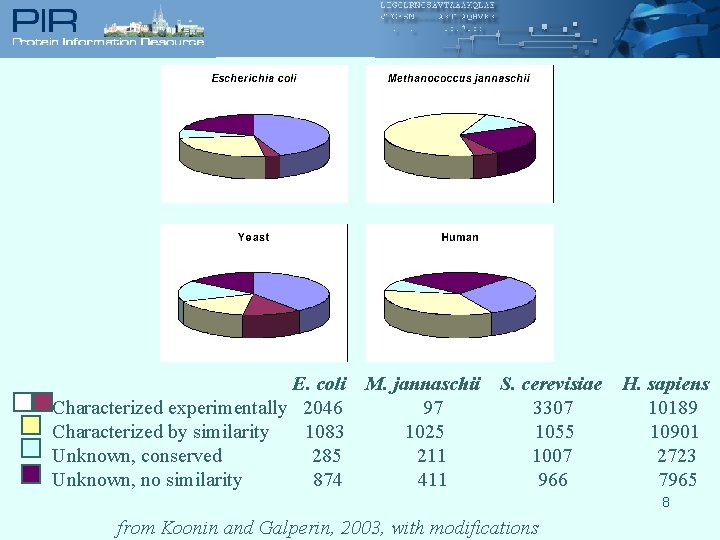
E. coli Characterized experimentally 2046 Characterized by similarity 1083 Unknown, conserved 285 Unknown, no similarity 874 M. jannaschii 97 1025 211 411 S. cerevisiae 3307 1055 1007 966 H. sapiens 10189 10901 2723 7965 8 from Koonin and Galperin, 2003, with modifications

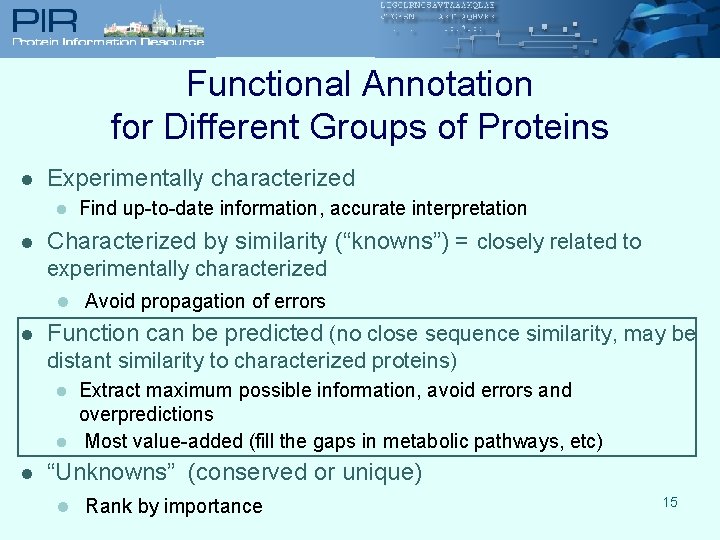
Functional Annotation for Different Groups of Proteins l Experimentally characterized l l Find up-to-date information, accurate interpretation Characterized by similarity (“knowns”) = closely related to experimentally characterized l Avoid propagation of errors l Function can be predicted (no close sequence similarity, may be distant similarity to characterized proteins) l l l Extract maximum possible information, avoid errors and overpredictions Most value-added (fill the gaps in metabolic pathways, etc) “Unknowns” (conserved or unique) l Rank by importance 9

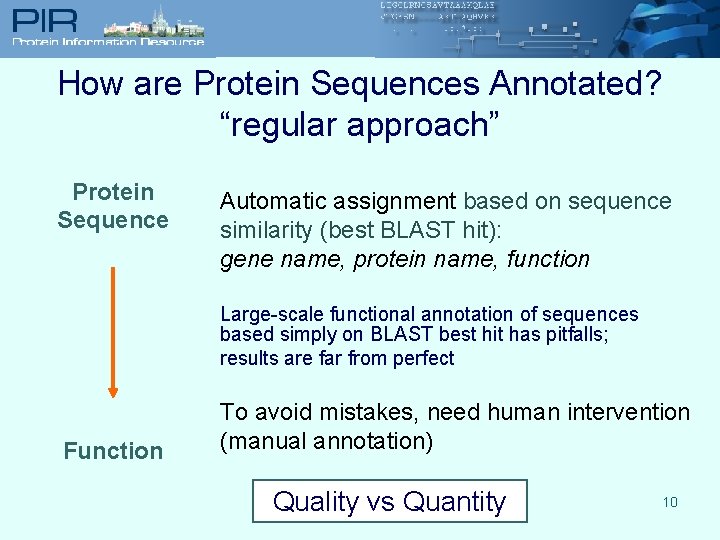
How are Protein Sequences Annotated? “regular approach” Protein Sequence Automatic assignment based on sequence similarity (best BLAST hit): gene name, protein name, function Large-scale functional annotation of sequences based simply on BLAST best hit has pitfalls; results are far from perfect Function To avoid mistakes, need human intervention (manual annotation) Quality vs Quantity 10

Functional Annotation for Different Groups of Proteins l Experimentally characterized l l Find up-to-date information, accurate interpretation Characterized by similarity (“knowns”) = closely related to experimentally characterized l Avoid propagation of errors l Function can be predicted (no close sequence similarity, may be distant similarity to characterized proteins) l l l Extract maximum possible information, avoid errors and overpredictions Most value-added (fill the gaps in metabolic pathways, etc) “Unknowns” (conserved or unique) l Rank by importance 11

Problems in Functional Assignments for “Knowns” l l Misinterpreted experimental results (e. g. suppressors, cofactors) Biologically senseless annotations Arabidopsis: separation anxiety protein-like Helicobacter: brute force protein Methanococcus: centromere-binding protein Plasmodium: frameshift l l - “Goofy” mistakes of sequence comparison (e. g. abc 1/ABC) Multi-domain organization of proteins Low sequence complexity (coiled-coil, transmembrane, nonglobular regions) Enzyme evolution: Divergence in sequence and function (minor mutation in active 12 site) Non-orthologous gene displacement: Convergent evolution

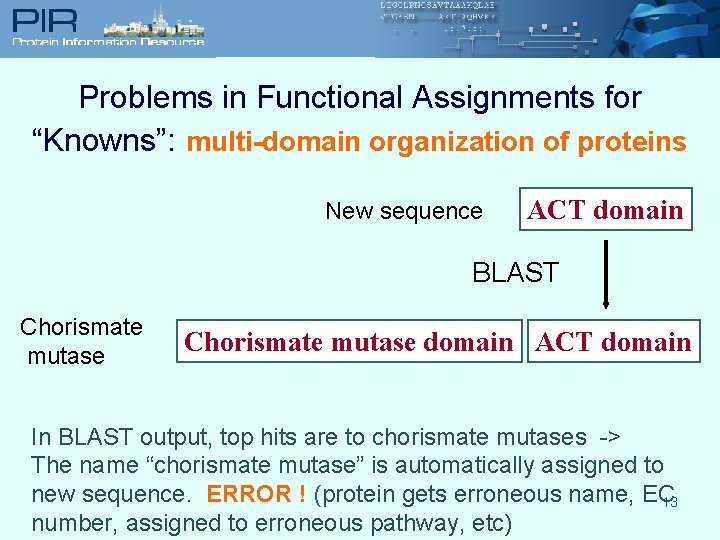
Problems in Functional Assignments for “Knowns”: multi-domain organization of proteins New sequence ACT domain BLAST Chorismate mutase domain ACT domain In BLAST output, top hits are to chorismate mutases -> The name “chorismate mutase” is automatically assigned to new sequence. ERROR ! (protein gets erroneous name, EC 13 number, assigned to erroneous pathway, etc)

Problems in Functional Assignments for “Knowns” Previous low quality annotations lead to propagation of mistakes 14

Functional Annotation for Different Groups of Proteins l Experimentally characterized l l Find up-to-date information, accurate interpretation Characterized by similarity (“knowns”) = closely related to experimentally characterized l Avoid propagation of errors l Function can be predicted (no close sequence similarity, may be distant similarity to characterized proteins) l l l Extract maximum possible information, avoid errors and overpredictions Most value-added (fill the gaps in metabolic pathways, etc) “Unknowns” (conserved or unique) l Rank by importance 15

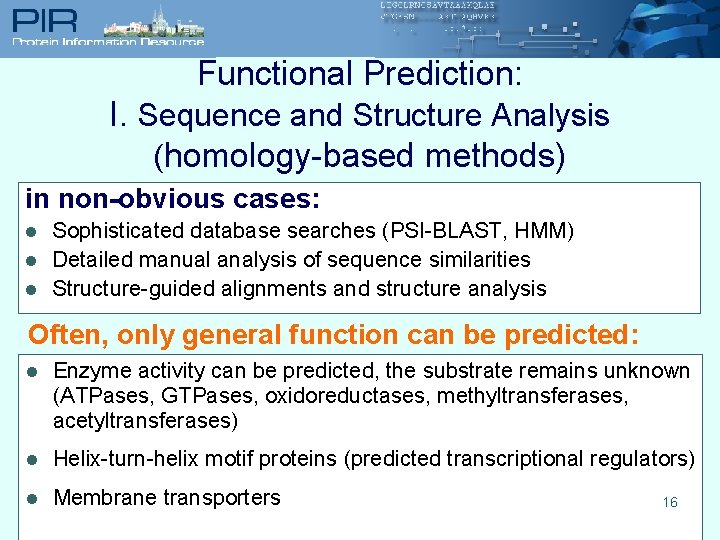
Functional Prediction: I. Sequence and Structure Analysis (homology-based methods) in non-obvious cases: l l l Sophisticated database searches (PSI-BLAST, HMM) Detailed manual analysis of sequence similarities Structure-guided alignments and structure analysis Often, only general function can be predicted: l Enzyme activity can be predicted, the substrate remains unknown (ATPases, GTPases, oxidoreductases, methyltransferases, acetyltransferases) l Helix-turn-helix motif proteins (predicted transcriptional regulators) l Membrane transporters 16

Using Sequence Analysis: Hints l Proteins (domains) with different 3 D folds are not homologous (unrelated by origin). Proteins with similar 3 D folds are usually (but not always) homologous l Those amino acids that are conserved in divergent proteins within a (super)family are likely to be functionally important (catalytic or binding sites, ect). l Reaction chemistry often remains conserved even when sequence diverges almost beyond recognition 17

Using Sequence Analysis: Hints l Prediction of 3 D fold (if distant homologs have known structures!) and of general biochemical function is much easier than prediction of exact biological function l Sequence analysis complements structural comparisons and can greatly benefit from them l Comparative analysis allows us to find subtle sequence similarities in proteins that would not have been noticed otherwise 18 Credit: Dr. M. Galperin, NCBI

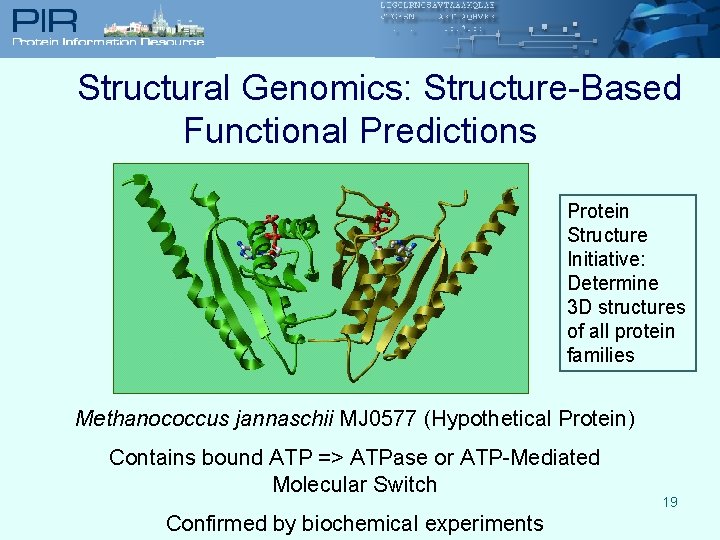
Structural Genomics: Structure-Based Functional Predictions Protein Structure Initiative: Determine 3 D structures of all protein families Methanococcus jannaschii MJ 0577 (Hypothetical Protein) Contains bound ATP => ATPase or ATP-Mediated Molecular Switch Confirmed by biochemical experiments 19

Crystal Structure is Not a Function! 20 Credit: Dr. M. Galperin, NCBI

Functional Prediction: II. Computational Analysis Beyond Homology l Phylogenetic distribution (comparative genomics) l l l Wide - most likely essential Narrow - probably clade-specific Patchy - most intriguing Clues: specific to niche, pathway type Domain association – “Rosetta Stone” Genome context (gene neighborhood, operon organization) 21

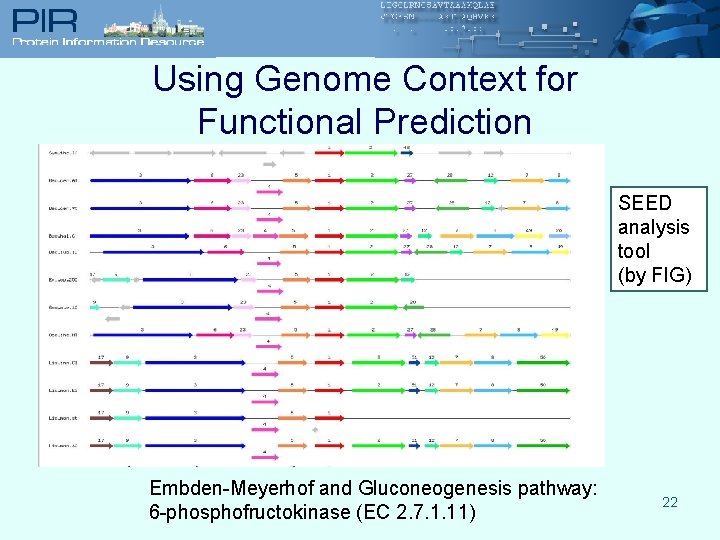
Using Genome Context for Functional Prediction SEED analysis tool (by FIG) Embden-Meyerhof and Gluconeogenesis pathway: 6 -phosphofructokinase (EC 2. 7. 1. 11) 22

Functional Prediction: Problem Areas l l Identification of protein-coding regions Delineation of potential function(s) for distant paralogs Identification of domains in the absence of close homologs Analysis of proteins with low sequence complexity 23

What to do with a new protein sequence Basic: - Domain analysis (SMART = most sensitive; PFAM, CDD) - BLAST - Curated protein family databases (PIRSF, Inter. Pro, COGs) - Literature (Pub. Med) from links from individual entries on BLAST output (look for Swiss. Prot entries first) l l - If not sufficient: PSI-BLAST Refined Pub. Med search using gene/protein names, synonyms, function and other terms you found Genome neighborhood (prokaryotes) Advanced: Multiple sequence alignments (manual) Structure-guided alignments and structure analysis - Phylogenetic tree reconstruction 24

Case Study: Prediction Verified: GGDEF domain l l l Proteins containing this domain: Caulobacter crescentus Ple. D controls swarmer cell - stalk cell transition (Hecht and Newton, 1995). In Rhizobium leguminosarum, Acetobacter xylinum, required for cellulose biosynthesis (regulation) Predicted to be involved in signal transduction because it is found in fusions with other signaling domains (receiver, etc) In Acetobacter xylinum, cyclic di-GMP is a specific nucleotide regulator of cellulose synthase (signalling molecule). Multidomain protein with GGDEF domain was shown to have diguanylate cyclase activity (Tal et al. , 1998) Detailed sequence analysis tentatively predicts GGDEF to be a diguanylate cyclase domain (Pei and Grishin, 2001) Complementation experiments prove diguanylate cyclase activity 25 of GGDEF (Ausmees et al. , 2001)

The Need for Classification Problem: l l Most new protein sequences come from genome sequencing projects Many have unknown functions Large-scale functional annotation of these sequences based simply on BLAST best hit has pitfalls; results are far from perfect Manual annotation of individual proteins is not efficient Solution: l l Highly curated annotated protein classification system Automatic annotation of sequences based on protein families Facilitates: l l Automatic annotation of sequences based on protein families Systematic correction of annotation errors Protein name standardization Functional predictions for uncharacterized proteins 26 This all works only if the system is optimized for annotation

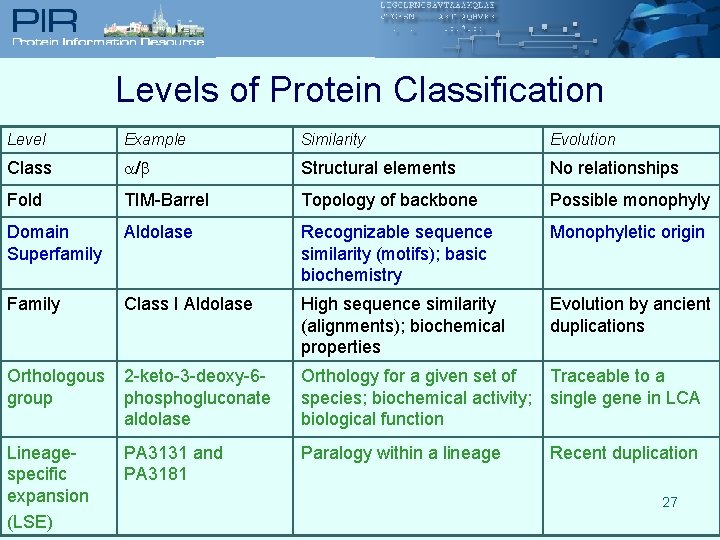
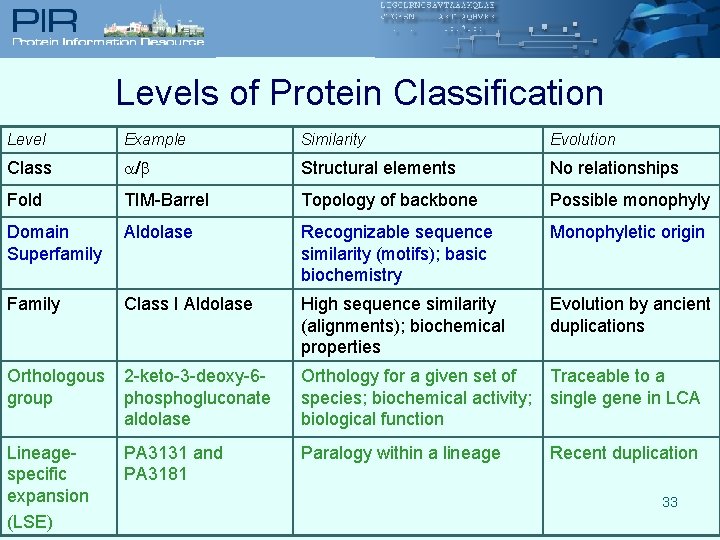
Levels of Protein Classification Level Example Similarity Evolution Class / Structural elements No relationships Fold TIM-Barrel Topology of backbone Possible monophyly Domain Superfamily Aldolase Recognizable sequence similarity (motifs); basic biochemistry Monophyletic origin Family Class I Aldolase High sequence similarity (alignments); biochemical properties Evolution by ancient duplications Orthologous group 2 -keto-3 -deoxy-6 phosphogluconate aldolase Orthology for a given set of species; biochemical activity; biological function Traceable to a single gene in LCA Lineagespecific expansion (LSE) PA 3131 and PA 3181 Paralogy within a lineage Recent duplication 27

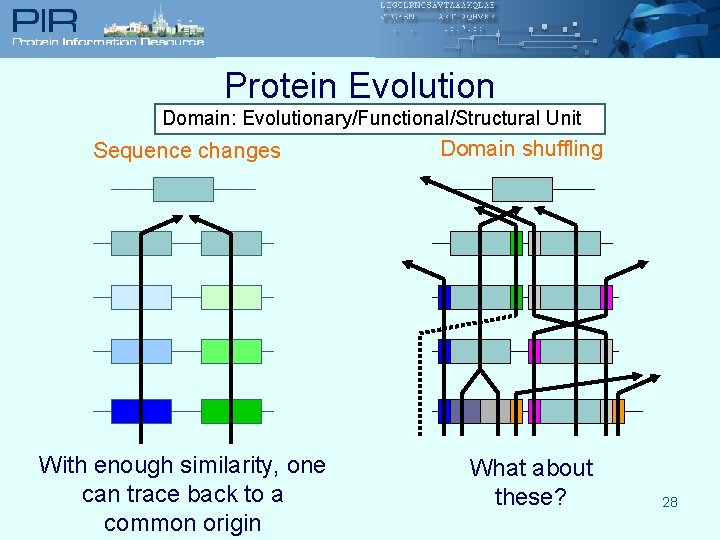
Protein Evolution Domain: Evolutionary/Functional/Structural Unit Sequence changes With enough similarity, one can trace back to a common origin Domain shuffling What about these? 28

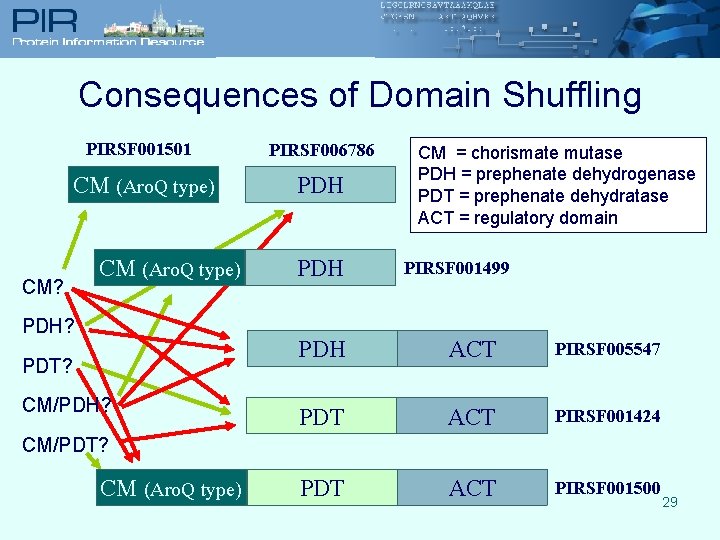
Consequences of Domain Shuffling PIRSF 001501 PIRSF 006786 CM (Aro. Q type) PDH CM? CM (Aro. Q type) PDH? PDT? CM/PDH? PDH CM = chorismate mutase PDH = prephenate dehydrogenase PDT = prephenate dehydratase ACT = regulatory domain PIRSF 001499 PDH ACT PIRSF 005547 PDT ACT PIRSF 001424 PDT ACT PIRSF 001500 CM/PDT? CM (Aro. Q type) 29

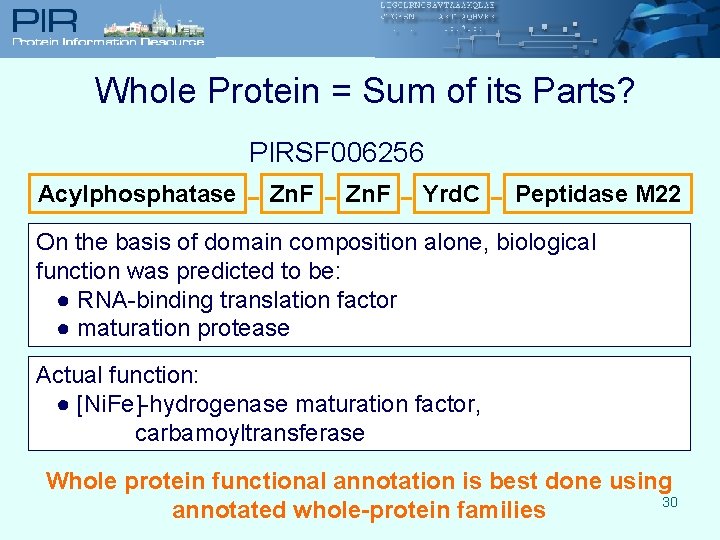
Whole Protein = Sum of its Parts? PIRSF 006256 Acylphosphatase - Zn. F - Yrd. C - Peptidase M 22 On the basis of domain composition alone, biological function was predicted to be: ● RNA-binding translation factor ● maturation protease Actual function: ● [Ni. Fe]-hydrogenase maturation factor, carbamoyltransferase Whole protein functional annotation is best done using 30 annotated whole-protein families

Practical classification of proteins: setting realistic goals We strive to reconstruct the natural classification of proteins to the fullest possible extent BUT Domain shuffling rapidly degrades the continuity in the protein structure (faster than sequence divergence degrades similarity) THUS The further we extend the classification, the finer is the domain structure we need to consider SO We need to compromise between the depth of analysis and protein integrity OR … 31 Credit: Dr. Y. Wolf, NCBI

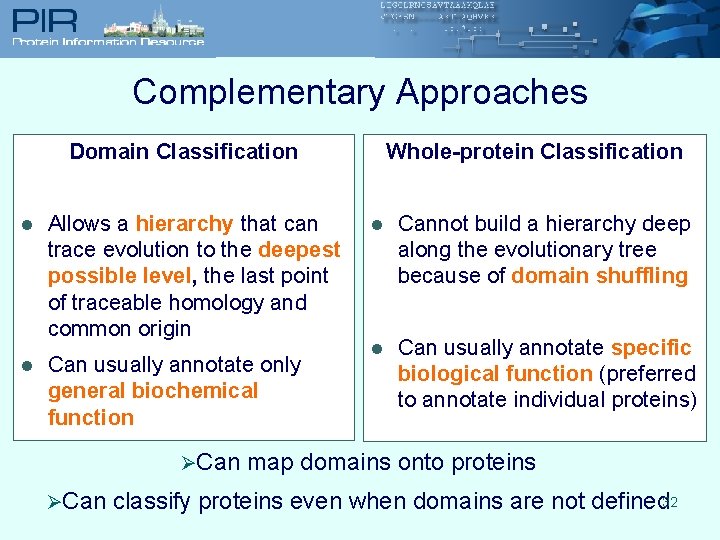
Complementary Approaches Whole-protein Classification Domain Classification l l Allows a hierarchy that can trace evolution to the deepest possible level, the last point of traceable homology and common origin Can usually annotate only general biochemical function ØCan l Cannot build a hierarchy deep along the evolutionary tree because of domain shuffling l Can usually annotate specific biological function (preferred to annotate individual proteins) map domains onto proteins classify proteins even when domains are not defined 32

Levels of Protein Classification Level Example Similarity Evolution Class / Structural elements No relationships Fold TIM-Barrel Topology of backbone Possible monophyly Domain Superfamily Aldolase Recognizable sequence similarity (motifs); basic biochemistry Monophyletic origin Family Class I Aldolase High sequence similarity (alignments); biochemical properties Evolution by ancient duplications Orthologous group 2 -keto-3 -deoxy-6 phosphogluconate aldolase Orthology for a given set of species; biochemical activity; biological function Traceable to a single gene in LCA Lineagespecific expansion (LSE) PA 3131 and PA 3181 Paralogy within a lineage Recent duplication 33

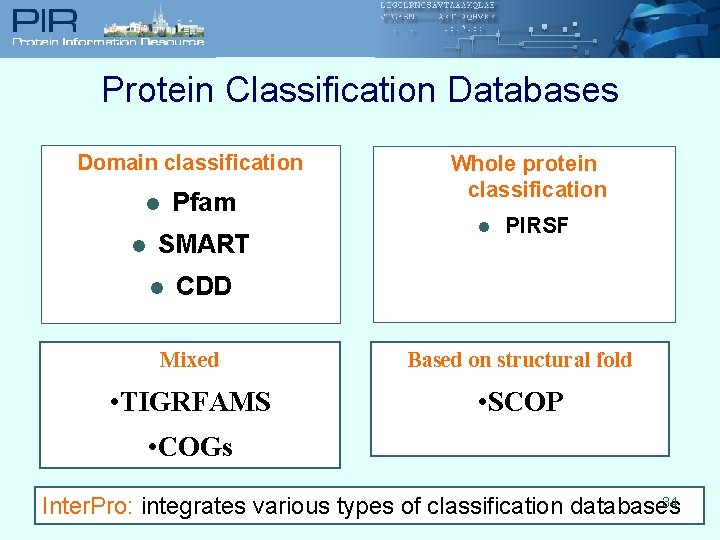
Protein Classification Databases Domain classification Pfam l l SMART l Whole protein classification l PIRSF CDD Mixed Based on structural fold • TIGRFAMS • SCOP • COGs 34 Inter. Pro: integrates various types of classification databases

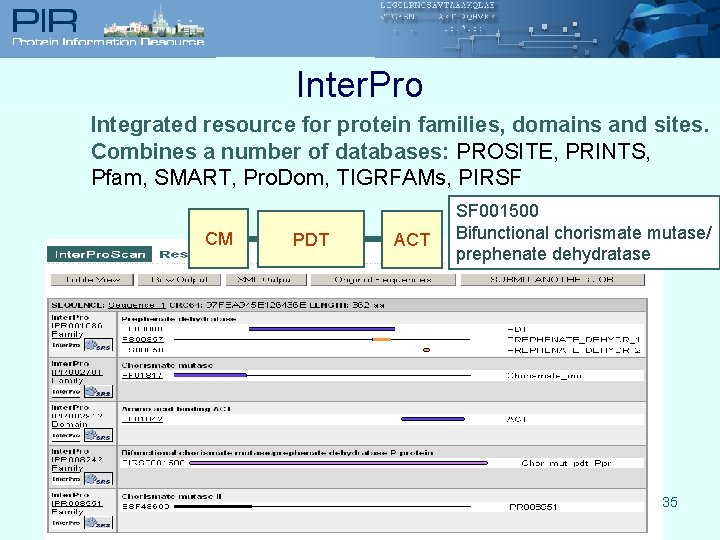
Inter. Pro Integrated resource for protein families, domains and sites. Combines a number of databases: PROSITE, PRINTS, Pfam, SMART, Pro. Dom, TIGRFAMs, PIRSF CM PDT ACT SF 001500 Bifunctional chorismate mutase/ prephenate dehydratase 35

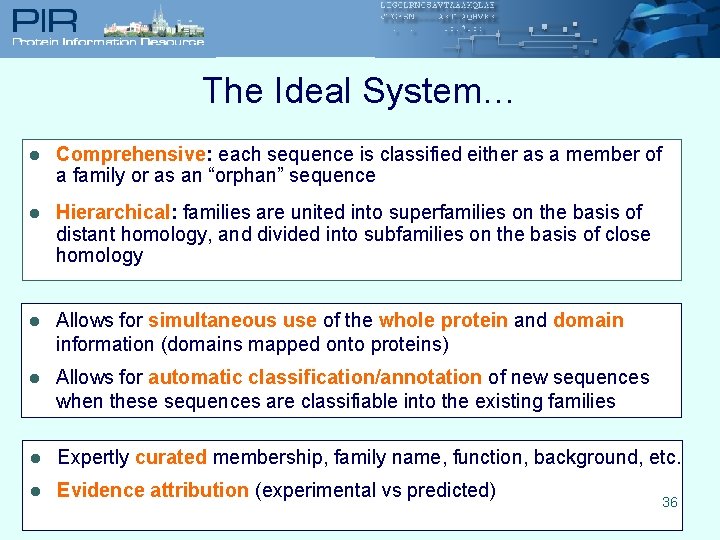
The Ideal System… l Comprehensive: each sequence is classified either as a member of a family or as an “orphan” sequence l Hierarchical: families are united into superfamilies on the basis of distant homology, and divided into subfamilies on the basis of close homology l Allows for simultaneous use of the whole protein and domain information (domains mapped onto proteins) l Allows for automatic classification/annotation of new sequences when these sequences are classifiable into the existing families l Expertly curated membership, family name, function, background, etc. l Evidence attribution (experimental vs predicted) 36

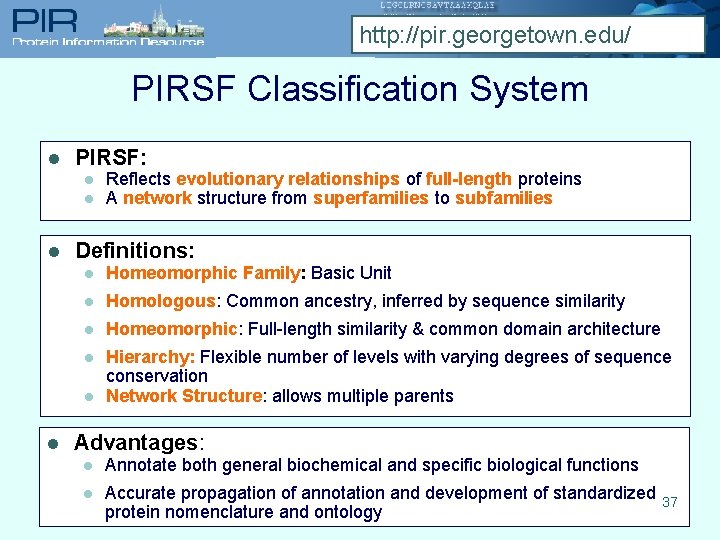
http: //pir. georgetown. edu/ PIRSF Classification System l PIRSF: l l l Definitions: l Homeomorphic Family: Basic Unit l Homologous: Common ancestry, inferred by sequence similarity l Homeomorphic: Full-length similarity & common domain architecture l Hierarchy: Flexible number of levels with varying degrees of sequence conservation Network Structure: allows multiple parents l l Reflects evolutionary relationships of full-length proteins A network structure from superfamilies to subfamilies Advantages: l Annotate both general biochemical and specific biological functions l Accurate propagation of annotation and development of standardized 37 protein nomenclature and ontology

PIRSF Classification System A protein may be assigned to only one homeomorphic family, which may have zero or more child nodes and zero or more parent nodes. Each homeomorphic family may have as many domain superfamily parents as its members have domains. 38

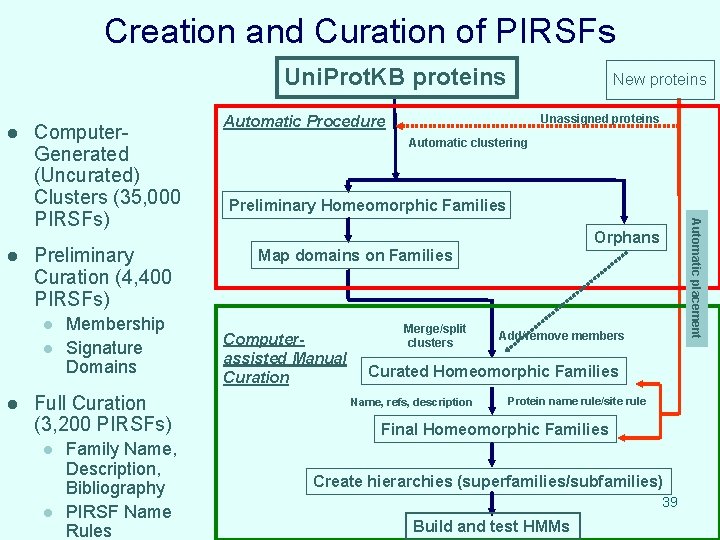
Creation and Curation of PIRSFs Uni. Prot. KB proteins l Preliminary Curation (4, 400 PIRSFs) l l l Membership Signature Domains Full Curation (3, 200 PIRSFs) l l Family Name, Description, Bibliography PIRSF Name Rules Unassigned proteins Automatic Procedure Automatic clustering Preliminary Homeomorphic Families Orphans Map domains on Families Computerassisted Manual Curation Merge/split clusters Automatic placement l Computer. Generated (Uncurated) Clusters (35, 000 PIRSFs) New proteins Add/remove members Curated Homeomorphic Families Name, refs, description Protein name rule/site rule Final Homeomorphic Families Create hierarchies (superfamilies/subfamilies) 39 Build and test HMMs

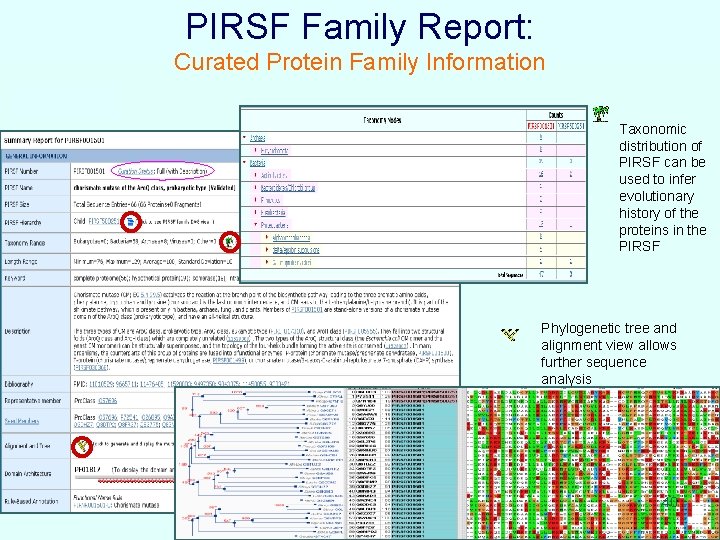
PIRSF Family Report: Curated Protein Family Information Taxonomic distribution of PIRSF can be used to infer evolutionary history of the proteins in the PIRSF Phylogenetic tree and alignment view allows further sequence analysis 40

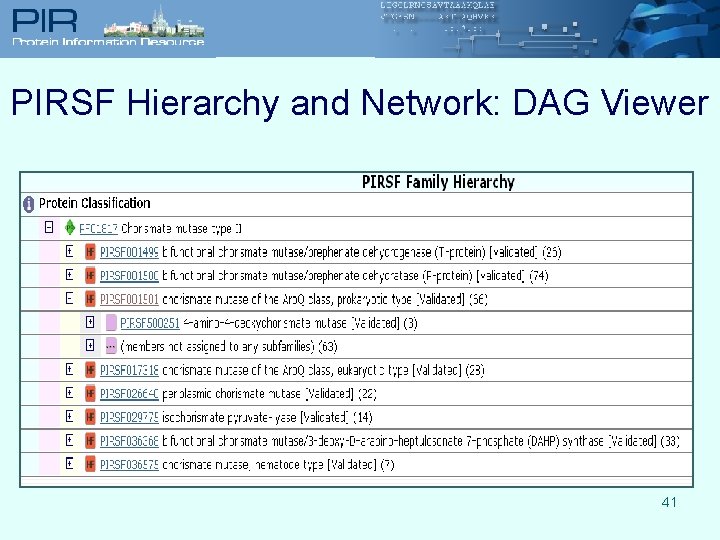
PIRSF Hierarchy and Network: DAG Viewer 41

PIRSF Family Report (II) Integrated value added information from other databases Mapping to other protein classification databases 42

PIRSF Protein Classification: Platform for Protein Analysis and Annotation l l Matching a protein sequence to a curated protein family rather than searching against a protein database Provides value-added information by expert curators, e. g. , annotation of uncharacterized hypothetical proteins (functional predictions) Improves automatic annotation quality Serves as a protein analysis platform for broad range of users 43

Family-Driven Protein Annotation Objective: Optimize for protein annotation l PIRSF Classification Name l l l Hierarchy l l Subfamilies increase specificity (kinase -> sugar kinase -> hexokinase) Name Rules l l Reflects the function when possible Indicates the maximum specificity that still describes the entire group Standardized format Name tags: validated, tentative, predicted, functionally heterogeneous Define conditions under which names propagate to individual proteins Enable further specificity based on taxonomy or motifs Names adhere to Swiss-Prot conventions (though we may make suggestions for improvement) Site Rules l Define conditions under which features propagate to individual proteins 44

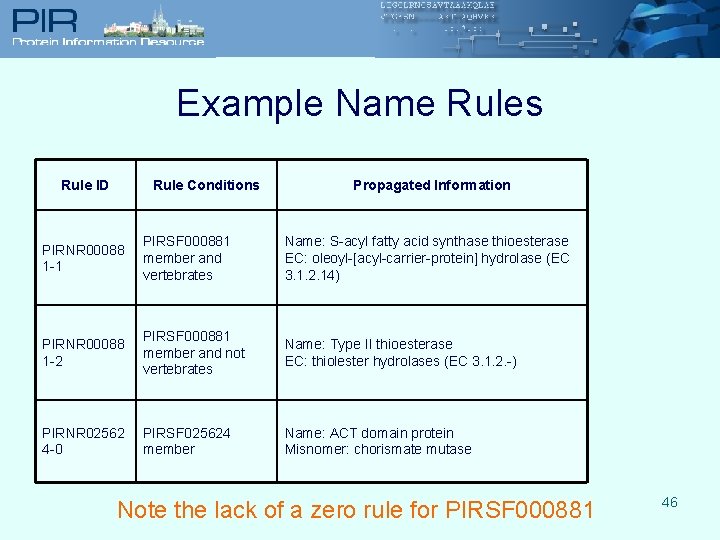
PIR Name Rules l Account for functional variations within one PIRSF, including: l l l Lack of active site residues necessary for enzymatic activity Certain activities relevant only to one part of the taxonomic tree Evolutionarily-related proteins whose biochemical activities are known to differ Monitor such variables to ensure accurate propagation l Propagate other properties that describe function: EC, GO terms, misnomer info, pathway l Name Rule types: l “Zero” Rule l l l Default rule (only condition is membership in the appropriate family) Information is suitable for every member “Higher-Order” Rule l l Has requirements in addition to membership 45 Can have multiple rules that may or may not have mutually exclusive conditions

Example Name Rules Rule ID Rule Conditions Propagated Information PIRNR 00088 1 -1 PIRSF 000881 member and vertebrates Name: S-acyl fatty acid synthase thioesterase EC: oleoyl-[acyl-carrier-protein] hydrolase (EC 3. 1. 2. 14) PIRNR 00088 1 -2 PIRSF 000881 member and not vertebrates Name: Type II thioesterase EC: thiolester hydrolases (EC 3. 1. 2. -) PIRNR 02562 4 -0 PIRSF 025624 member Name: ACT domain protein Misnomer: chorismate mutase Note the lack of a zero rule for PIRSF 000881 46

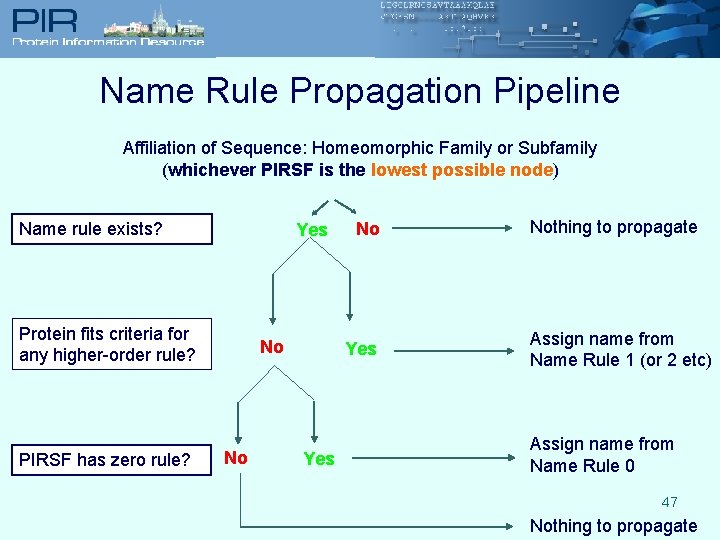
Name Rule Propagation Pipeline Affiliation of Sequence: Homeomorphic Family or Subfamily (whichever PIRSF is the lowest possible node) Name rule exists? Yes Protein fits criteria for any higher-order rule? PIRSF has zero rule? No No No Yes Nothing to propagate Assign name from Name Rule 1 (or 2 etc) Assign name from Name Rule 0 47 Nothing to propagate

Name Rule in Action at Uni. Prot Current: • Automatic annotations (AA) are in a separate field • AA only visible from www. ebi. uniprot. org Future: • Automatic name annotations will become DE line if DE line will improve as a result • AA will be visible from all consortium-hosted web sites 48

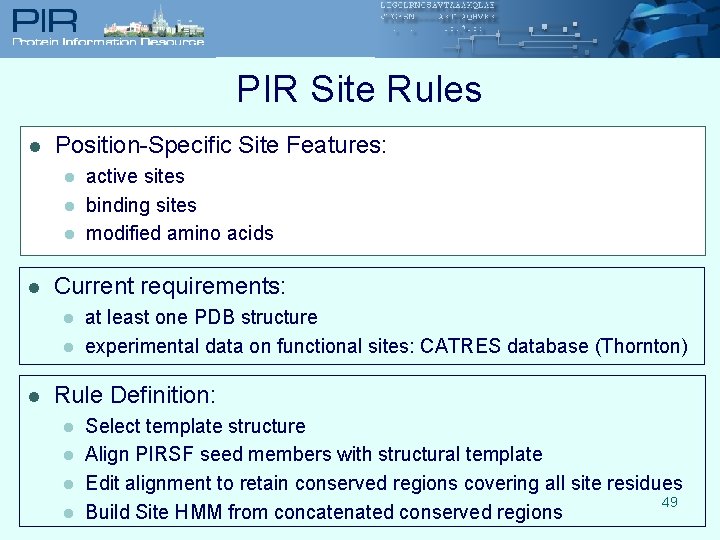
PIR Site Rules l Position-Specific Site Features: l l Current requirements: l l l active sites binding sites modified amino acids at least one PDB structure experimental data on functional sites: CATRES database (Thornton) Rule Definition: l l Select template structure Align PIRSF seed members with structural template Edit alignment to retain conserved regions covering all site residues 49 Build Site HMM from concatenated conserved regions

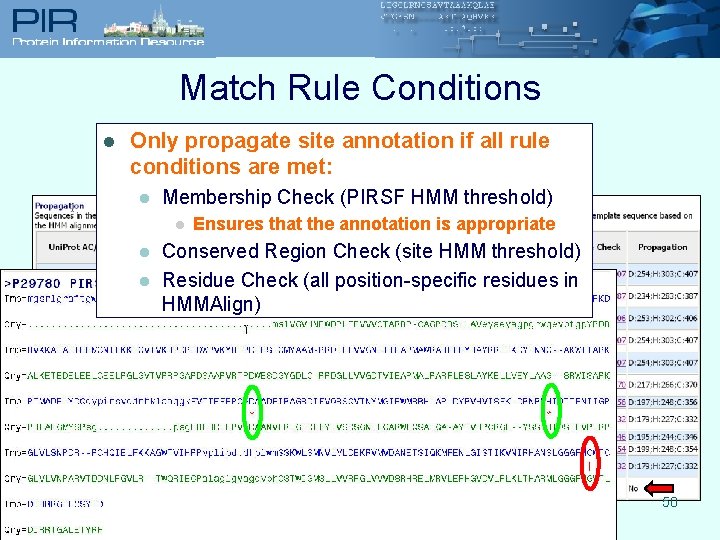
Match Rule Conditions l Only propagate site annotation if all rule conditions are met: l Membership Check (PIRSF HMM threshold) l l l Ensures that the annotation is appropriate Conserved Region Check (site HMM threshold) Residue Check (all position-specific residues in HMMAlign) 50

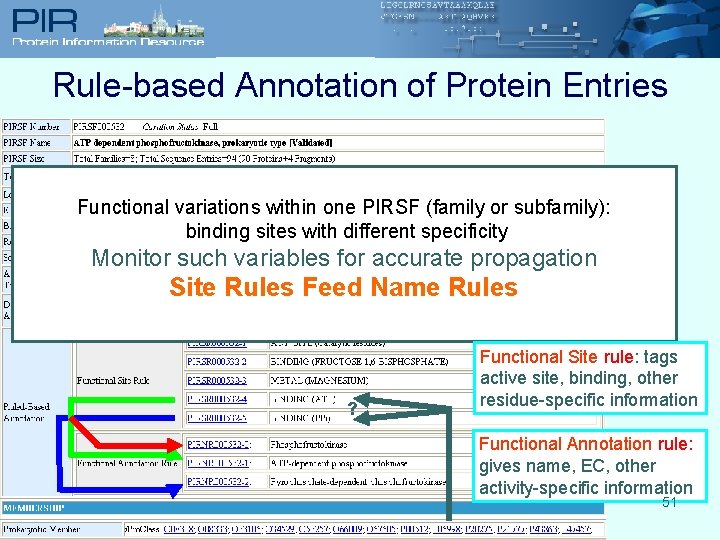
Rule-based Annotation of Protein Entries Functional variations within one PIRSF (family or subfamily): binding sites with different specificity Monitor such variables for accurate propagation Site Rules Feed Name Rules ? Functional Site rule: tags active site, binding, other residue-specific information Functional Annotation rule: gives name, EC, other activity-specific information 51

Problem: l l l Overview Most new protein sequences come from genome sequencing projects Many have unknown functions Large-scale functional annotation of these sequences based simply on BLAST best hit has pitfalls; results are far from perfect Functional Analysis of Protein Sequences: l l Homology-based (sequence analysis, structure analysis) Non-homology (genome context, phylogenetic distribution) Solution for Large-scale Annotation: l l Highly curated annotated protein classification system Automatic annotation of sequences based on protein families Facilitates: l l Automatic annotation of sequences based on protein families Systematic correction of annotation errors Name standardization in Uni. Prot Functional predictions for uncharacterized proteins 52

Impact of Protein Bioinformatics and Genomics l l l Single protein level l Discovery of new enzymes and superfamilies l Prediction of active sites and 3 D structures Pathway level l Identification of “missing” enzymes l Prediction of alternative enzyme forms l Identification of potential drug targets Cellular metabolism level l Multisubunit protein systems l Membrane energy transducers l Cellular signaling systems 53

PIR Team l l l Dr. Cathy Wu, Director Protein Science team l Dr. Darren Natale (lead) l Dr. Cecilia Arighi l Dr. Winona Barker l Dr. Zhang-zhi Hu l Dr. Raja Mazumder Bioinformatics team l Dr. Hongzhan Huang (lead) l Dr. Leslie Arminski l Dr. Hsing-Kuo Hua l Dr. Robel Kahsay Students l Natalia Petrova Uni. Prot Collaborators l Dr. Rolf Apweiler (EBI) Dr. Peter Mc. Garvey Dr. Anastasia Nikolskaya Dr. Sona Vasudevan Dr. CR Vinayaka Dr. Lai-Su Yeh Yongxing Chen, M. S. Baris Suzek, M. S. Xin Yuan, M. S. Jian Zhang, M. S. 54 Dr. Amos Bairoch (SIB)