Full name Denaturing Gradient Gel Electrophoresis Definition A


















- Slides: 27


머리말 ã ã ã Full name: - Denaturing Gradient Gel Electrophoresis Definition: - A molecular fingerprinting method that separates polymerase chain reaction (PCR)-generated DNA products Principles: - Generating templates of differing DNA sequence that represent many of the dominant microbial organisms - Separating PCR products based on sequence differences that results in differential denaturing characteristics of the DNA - Differing sequences denature at different denaturant concentrations slow movement diff position of band location - Each band theoretically representing a different bacterial population present in the community 2

머리말 - fingerprint similarity can be assessed to determine microbial structural differences between environments or among treatments - used to investigate broad phylogenies or specific target organisms such as pathogens or xenobiotics degraders 3

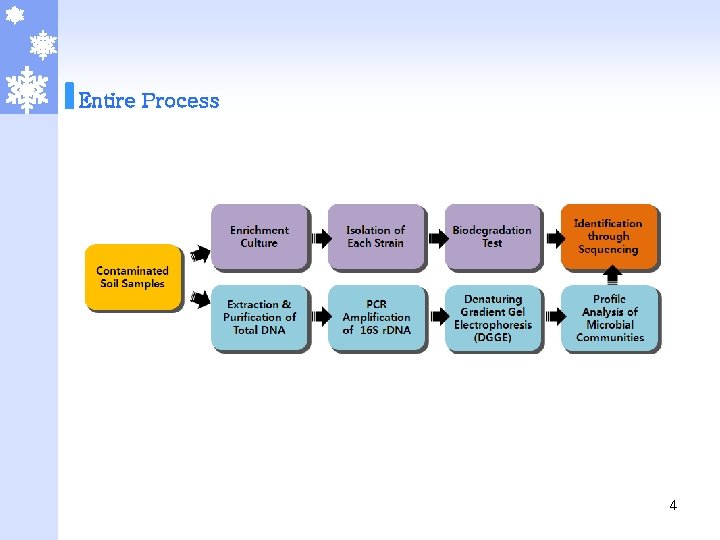
Entire Process 4


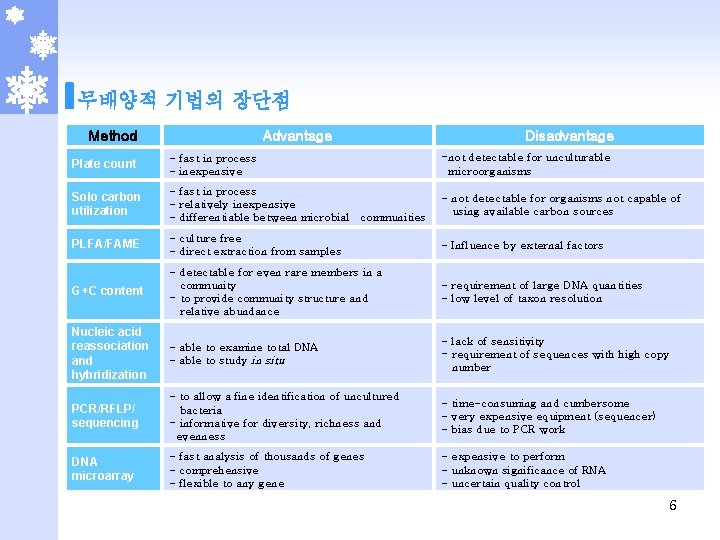
무배양적 기법의 장단점 Method Advantage Disadvantage Plate count - fast in process - inexpensive -not detectable for unculturable microorganisms Solo carbon utilization - fast in process - relatively inexpensive - differentiable between microbial communities - not detectable for organisms not capable of using available carbon sources PLFA/FAME - culture free - direct extraction from samples - Influence by external factors G+C content - detectable for even rare members in a community - to provide community structure and relative abundance - requirement of large DNA quantities - low level of taxon resolution Nucleic acid reassociation and hybridization - able to examine total DNA - able to study in situ - lack of sensitivity - requirement of sequences with high copy number PCR/RFLP/ sequencing - to allow a fine identification of uncultured bacteria - informative for diversity, richness and evenness - time-consuming and cumbersome - very expensive equipment (sequencer) - bias due to PCR work DNA microarray - fast analysis of thousands of genes - comprehensive - flexible to any gene - expensive to perform - unknown significance of RNA - uncertain quality control 6

무배양적 기법의 장단점 DGGE & TGGE - analysis of a large number of samples - to provide community structure and relative abundance - informative for the structure of active bacterial populations through RT-PCR - possible for taxonomic identification - easy to perform - less time-consuming - sensitive by sample handling - possible more than one stable form of ss. DNA - detectable for only most abundant species - not able to separate amplicons harboring different sequences - formation of chimaeric equences - differential PCR - not efficient separation beyond 500 base pairs - to migrate at the same position with two different sequences ARDRA - able to detect structural changes in microbial community - convenience (no more development) - easy to perform - less time-consuming - choice of suitable restriction enzymes - not always possible to make good resolution for high molecular fragments - formation of chimaeric equences - differential PCR T-RFLP - able to detect only the terminal fragments - easy to perform - less time-consuming - formation of chimaeric equences - differential PCR RISA - to provide a finer taxonomic identification - easy to perform - less time-consuming - not enough information on IGS sequence databank - not always possible to make good resolution for high molecular fragments - formation of chimaeric sequences - differential PCR RAPD - rapid and sensitive for revealing differences between similar complex genomes - easy to perform - less time-consuming - lack of reproducibility not able to provide phylogenetic information of chimaeric sequences differential PCR 7

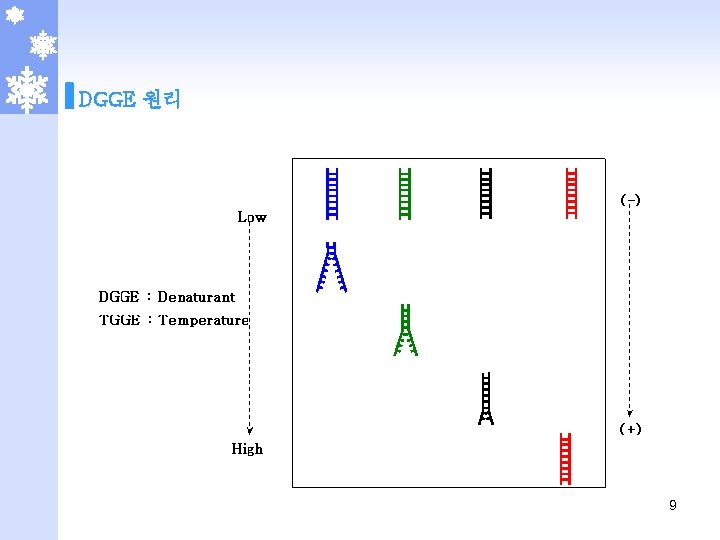
DGGE 원리 8

DGGE 원리 9

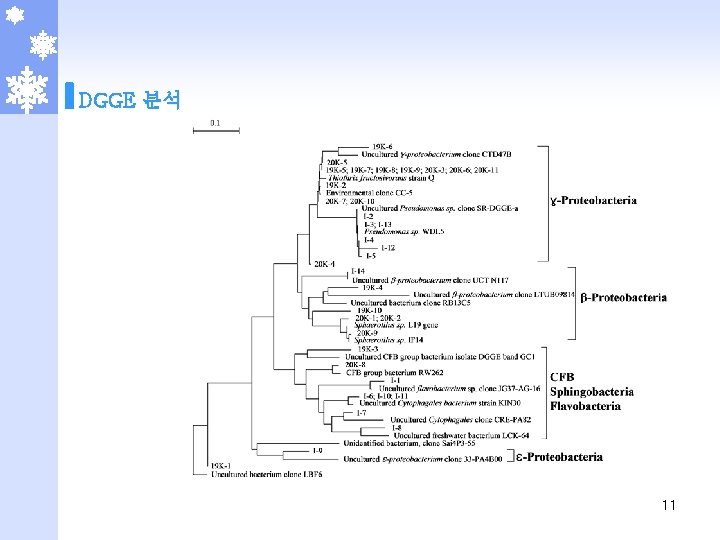
2. 본 론 DGGE 분석 OS PDR PDB PUR PUB UDB OS 100. 00 PDR 30. 43 100. 00 PDB 40. 00 41. 38 100. 00 PUR 30. 77 53. 85 40. 63 100. 00 PUB 37. 50 44. 44 57. 14 59. 26 100. 00 UDB 20. 69 36. 67 76. 92 45. 16 51. 72 100. 00 UUB 44. 44 39. 13 42. 31 50. 00 45. 83 42. 31 UUB 10 100. 00

DGGE 분석 11

Materials ã Plate sandwich materials - 16 x 16 cm glass plate - 16 x 14 cm glass plate - 2 - 1 mm spacers - 2 - plate clamps - Pouring stand - Foam gasket - Well comb - Spacing card 12

Materials ã ã ã Gel solutions - 40% Bis-Acrylamide: (37. 5: 1, acrylamide: bisacrylamide) - Deionized Formamide - Urea - 50 X TAE (20 m. M Tris-acetate, p. H 7. 4, 10 m. M sodium acetate, 0. 5 m. M Na-EDTA) - 10% Ammonium Persulfate Solution (APS, 0, 1 g in 1 ml of nanopure water. Make fresh when required or aliquot 0. 5 ml into 1. 5 ml microcentrifuge tubes and freeze until needed) - TEMED (N, N, N' -tetramethylenediamine) - Glycerol - Nanopure water Gradient wheel Gel. Star nucleic acid stain DCODE electrophoresis apparatus Power supply 13

Procedure A. Building the gel assembly 14

Procedure B. Pouring the gel 15

Procedure C. Running the gel Gels attached to the core assembly 16

Procedure D. Staining the gel 17



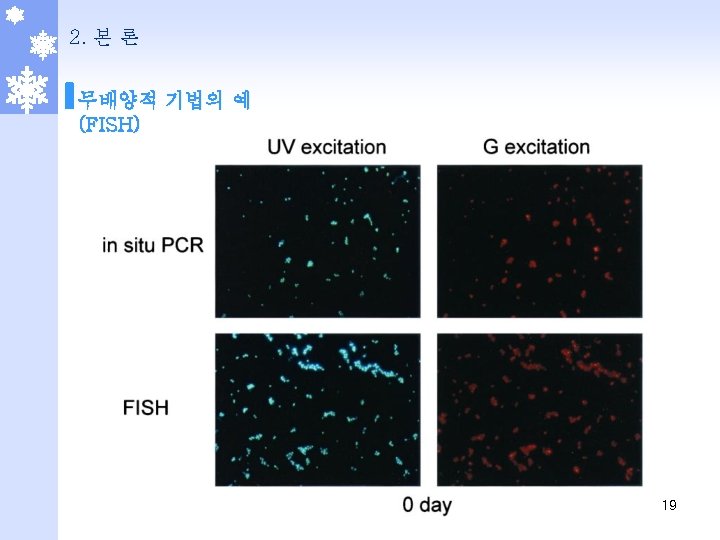
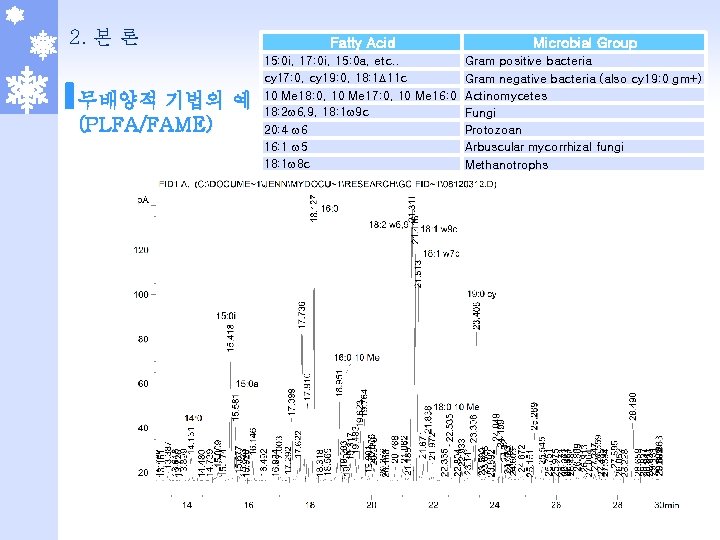
2. 본 론 무배양적 기법의 예 (PLFA/FAME) Fatty Acid Microbial Group 15: 0 i, 17: 0 i, 15: 0 a, etc. . Gram positive bacteria cy 17: 0, cy 19: 0, 18: 1 D 11 c Gram negative bacteria (also cy 19: 0 gm+) 10 Me 18: 0, 10 Me 17: 0, 10 Me 16: 0 Actinomycetes 18: 2 w 6, 9, 18: 1 w 9 c Fungi 20: 4 w 6 Protozoan 16: 1 w 5 Arbuscular mycorrhizal fungi 18: 1 w 8 c Methanotrophs 20


2. 본 론 Bacteria Methanogens 혐기성 세균의 계통발생적 분류 Sulfate-reducers Phylogenetic Group Species Euryarchaeotes Methanobacterium thermoautotrophicum 1) Methanobrevibacter arboriphilicus 1) Methanococcus voltae 1) Methanococcus termolithotrophicus 1) Methanosarcina barkeri 1) Euryarchaeotes Archaeoglobus sp. 4) δ-Proteobacteria Desulfoarculus sp. 4) Desulfobacterium sp. 4) Desulfobotulus sp. 4) Desulfobulbus sp. 3, 4) Desulfocapsa sp. 3) Desulfococcus sp 3, 4) Desulfomicrobium sp. 4) Desulfomonile sp. 4) Desulfonema sp. 4) Desulforhopalus sp. 3) Desulfosarcina sp. 3, 4) Desulfotomaculum sp. 4) Desulfovibrio sp. 4) Thermodesulfobacte ria Thermodesulfobacterium sp. 4) Crenarchaeota Sulfolobus acidocaldarius 6) Firmicutes Bacillus sp. 5) Clostridium sp. 6) Staphylococcus aureus 5) β-Proteobacteria Rhodoferax ferrireducens 7) Thiobacillus ferrooxidans 6) Thiobacillus thiooxidans 6) γ-Proteobacteria Acinetobacter calcoaceticus 5) Acinetobacter johnsonii 5) Pseudomonas fluorescens 5) Pseudomonas sp. 5, 6) Shewanella putrefaciens 5, 6) δ-Proteobacteria Desulfuromonas acetoxidans 5) Desulfovibrio sp. 6) Desulfovibrio vulgaris 6) Geobacter metallireducans 6, 7) Geobacter sp. 2, 6) Fe(III)-reducers 22

2. 본 론 혐기성 세균의 계통발생적 분류 Mn(IV)-reducers Firmicutes Bacillus sp. 5) Staphylococcus aureus 5) γ-Proteobacteria Acinetobacter calcoaceticus 5) Acinetobacter johnsonii 5) Pseudomonas fluorescens 5) Pseudomonas sp. 5, 6) Shewanella putrefaciens 5, 6) δ-Proteobacteria Desulfuromonas acetoxidans 5, 6) Geobacter metallireducans 5, 6) Geobacter sp. 2) Actinobacteria Corynebacterium diphtheriae 8) Corynebacterium efficiens 8) Corynebacterium glutamicum 8) Mycobacterium bovis 7) Mycobacterium tuberculosis 8) Rubrobacter xylanophilus 8) Streptomyces coelicolor 8) Crenarchaeotes Aeropyrum pernix 8) Pyrogaculum aerophilum 8) Euryarchaeotes Haloarcula marismoritui 8) Firmicutes Bacillus anthracis 8) Bacillus cereus 8) Bacillus licheniformis 8) Bacillus stearothermophilus 8) Bacillus subtilis 8) Geobacillus stearothermophilus 8) Lactobacillus plantarum 8) Selenomonas ruminantium 8) Staphylococcus aureus 8) Staphylococcus carnosus 8) Staphylococcus epidermidis 8) Nitrate-reducers 23

2. 본 론 �-Proteobacteria Azospirillum brasilense 8) Bradyrhizoium japonicum 8) Brucella suis 8) Brucella melitensis 8) Magnetospirillum magnetotacticum 8) Paracoccus denitrificans 8) Rhodobacter sphaeroides 8) Sinorhizobium meliloti 8) β-Proteobacteria Azoarcus sp. 2) Bordetella bronchiseptica 8) Bordetella parapertussis 8) Burkholderia cepacia 8) Burkhoderia fungorum 8) Burkholderia pseudomallei 8) Chromobacterium violaceum 8) Ralstonia metallisurans 8) Ralstonia solanacearum 8) Wautersia eutropha 8) γ-Proteobacteria Actinobacillus actinocydetemcomitans 8) Escherichia coli 8) Haemophilus ducreyi 8) Haemophilus influenzae 8) Halomonas halodenitrificans 8) Pasteurella multocida 8) Photobacterium profundum 8) Pseudomonas aeruginosa 8) Pseudomonas fluorescens 8) Psychrobacter sp. 8) Salmonella enterica 8) Salmonella typhimurium 8) Shewanella oneidensis 8) Shigella flexneri 8) δ-Proteobacteria Geobacter metallireducans 8) ε-Proteobacteria Campylobacter jejuni 8) Helicobacter hepticus 8) Sulfurospirillum barnesii 8) Wolinella succinogenes 8) Thermus thermophilus 8) 혐기성 세균의 계통발생적 분류 Nitrate-reducers 24

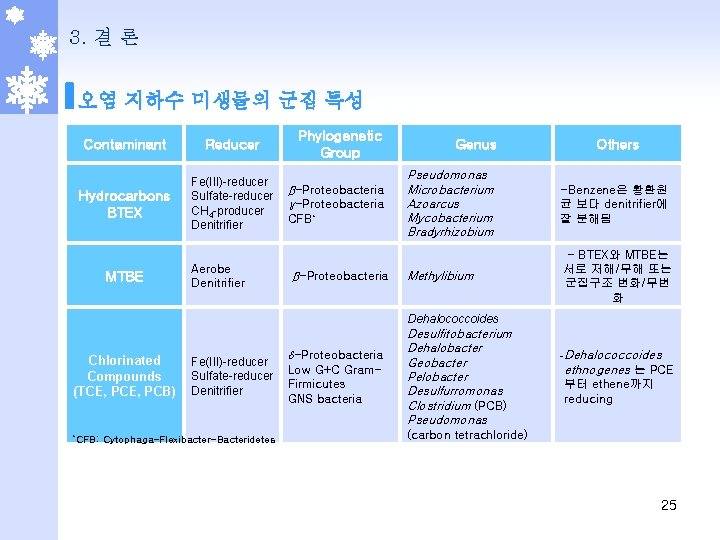
3. 결 론 오염 지하수 미생물의 군집 특성 Contaminant Reducer Hydrocarbons BTEX Fe(III)-reducer Sulfate-reducer CH 4 -producer Denitrifier MTBE Aerobe Denitrifier Phylogenetic Group β-Proteobacteria γ-Proteobacteria CFB* β-Proteobacteria Genus Others Pseudomonas Microbacterium Azoarcus Mycobacterium Bradyrhizobium -Benzene은 황환원 균 보다 denitrifier에 잘 분해됨 Methylibium - BTEX와 MTBE는 서로 저해/무해 또는 군집구조 변화/무변 화 Dehalococcoides Chlorinated Compounds (TCE, PCB) *CFB: Fe(III)-reducer Sulfate-reducer Denitrifier Cytophaga-Flexibacter-Bacteridetes -Proteobacteria Low G+C Gram. Firmicutes GNS bacteria Desulfitobacterium Dehalobacter Geobacter Pelobacter Desulfurromonas Clostridium (PCB) Pseudomonas -Dehalococcoides ethnogenes 는 PCE 부터 ethene까지 reducing (carbon tetrachloride) 25


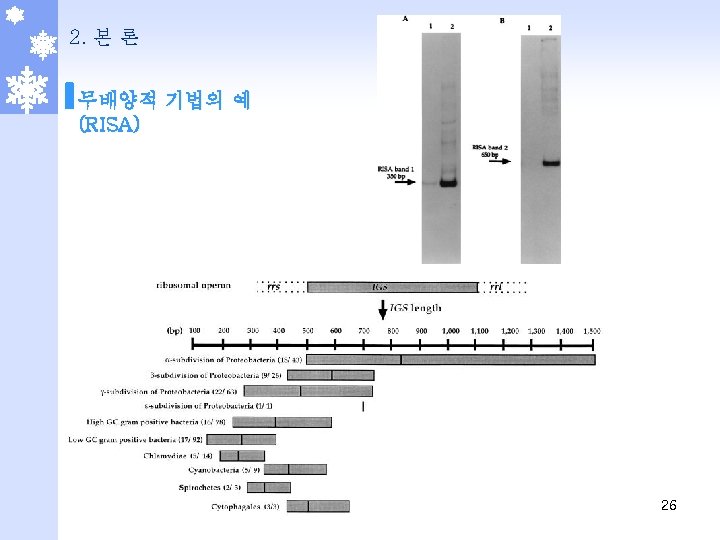
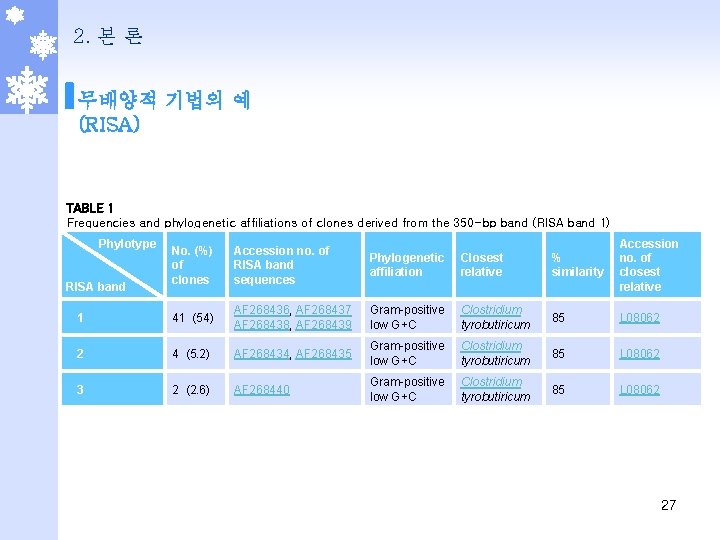
2. 본 론 무배양적 기법의 예 (RISA) TABLE 1 Frequencies and phylogenetic affiliations of clones derived from the 350 -bp band (RISA band 1) Phylotype No. (%) of clones Accession no. of RISA band sequences Phylogenetic affiliation Closest relative % similarity Accession no. of closest relative 1 41 (54) AF 268436, AF 268437 AF 268438, AF 268439 Gram-positive low G+C Clostridium tyrobutiricum 85 L 08062 2 4 (5. 2) AF 268434, AF 268435 Gram-positive low G+C Clostridium tyrobutiricum 85 L 08062 3 2 (2. 6) AF 268440 Gram-positive low G+C Clostridium tyrobutiricum 85 L 08062 RISA band 27