Fruit fly Drosophila melanogaster Fruit fly 180 Mb

Fruit fly Drosophila melanogaster

Fruit fly 180 Mb 13500 genes simple breeding in the lab small size easy to perform crossings short reproductive cycle (2 weeks) – for genetics 100 flies progeny genome sequenced in 2000 easy to obtain mutants, different mutants molecular biology - methods -2/3 human diseases -beginning: TH Morgan and chromosome theory -today: model for developmental biology and behavior molecular and cellular biology -genome not duplicated

Model organism from 1906 I. period 1910 -1940 -period of fast development of basic principles of classical genetics: chromosomal theory of inheritance nature of X linked inheritance and genetic maps genetics of chromosomal aberrations induction of genetic and chromosomal mutations mitotic recombination
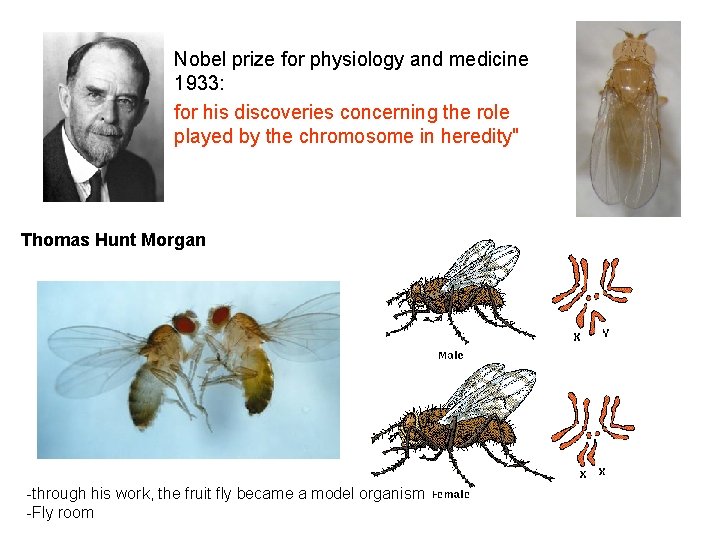
Nobel prize for physiology and medicine 1933: for his discoveries concerning the role played by the chromosome in heredity" Thomas Hunt Morgan -through his work, the fruit fly became a model organism -Fly room

Fly room Party in the Fly Room at Columbia on January 2, 1919. (L-R, back row): E. G. Anderson, A. Weinstein, S. C. Dellinger, C. B. Bridges, Pithecanthropus, H. J. Muller, T. H. Morgan, F. E. Lutz; (L-R, front row): F. Schrader, A. H. Sturtevant, A. F. Huettner, O. L. Mohr. Photo ID 1. 43 Thomas Hunt Morgan and his studends in the „Fly room”: Sturtevant, Morgan, Bridges, Muller
II. period 1940 -1968 investigations on microorganisms and phages III. period 1968 -2000 -development of analytic, instead of descriptive science considering development and behavior -technical advances in situ hybridization, cell transformation by P element, clonal analysis, discovery of strong chemical mutagens, molecular biology -fruit fly chosen as a model organism for the study of fundamental problems in biology (Genome Res. 15: 1661 -1667, 2005)

The Nobel Prize in Physiology or Medicine 1995 The Nobel Assembly at the Karolinska Institute in Stockholm, Sweden, has awarded the Nobel Prize in Physiology or Medicine for 1995 to Edward B. Lewis, Christiane Nüsslein-Volhard and Eric Wieschaus for their discoveries concerning "the genetic control of early embryonic development".

IV. period 2000„ genomic era” in 2000 DNA sequence of Drosophila Adams, M. D. et al. The genome sequence of Drosophila melanogaster. Science 287, 2185 -95 (2000).

- genetics and chromosomes of fruit fly - transposons – movable genetic elements - homeotic genes, transcription factors and development of fruit fly - today: different cellular and developmental processes - gene regulation
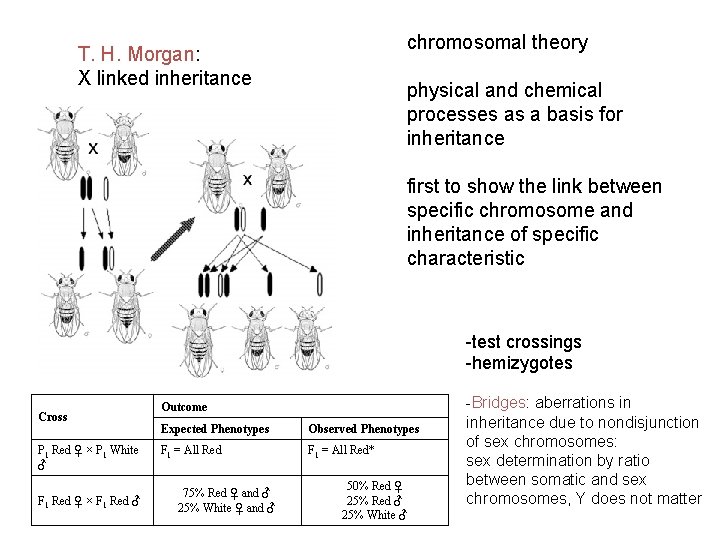
chromosomal theory T. H. Morgan: X linked inheritance physical and chemical processes as a basis for inheritance first to show the link between specific chromosome and inheritance of specific characteristic -test crossings -hemizygotes Cross P 1 Red ♀ × P 1 White ♂ F 1 Red ♀ × F 1 Red ♂ Outcome Expected Phenotypes Observed Phenotypes F 1 = All Red* 75% Red ♀ and ♂ 25% White ♀ and ♂ 50% Red ♀ 25% Red ♂ 25% White ♂ -Bridges: aberrations in inheritance due to nondisjunction of sex chromosomes: sex determination by ratio between somatic and sex chromosomes, Y does not matter
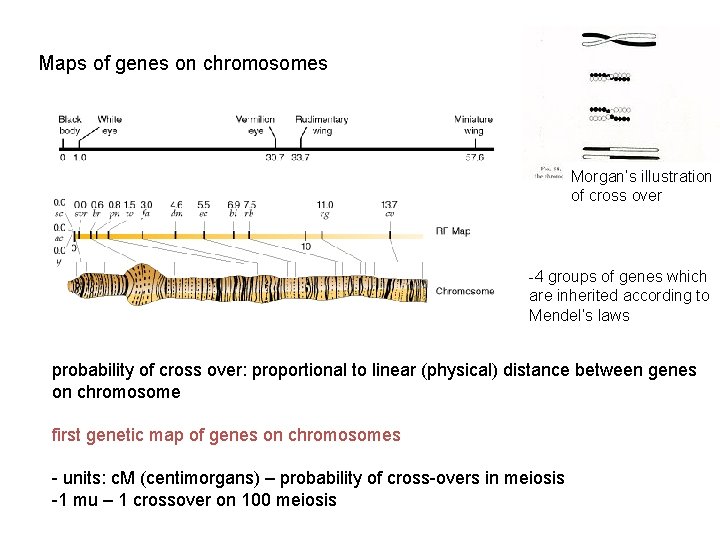
Maps of genes on chromosomes Morgan’s illustration of cross over -4 groups of genes which are inherited according to Mendel’s laws probability of cross over: proportional to linear (physical) distance between genes on chromosome first genetic map of genes on chromosomes - units: c. M (centimorgans) – probability of cross-overs in meiosis -1 mu – 1 crossover on 100 meiosis
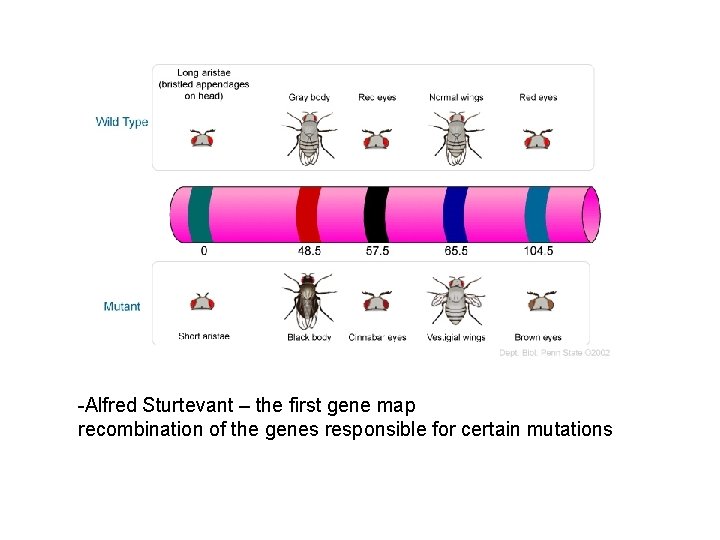
-Alfred Sturtevant – the first gene map recombination of the genes responsible for certain mutations
Hermann J. Muller -1946: Nobel prize for medicine -fruit fly mutagenesis by X ray irradiation induced mutations -recessive lethal phenotype x irradiation increases the rate of mutations cross over only in females (easier genetic manipulations and screening) - sometimes one feature depends on more genes and their -
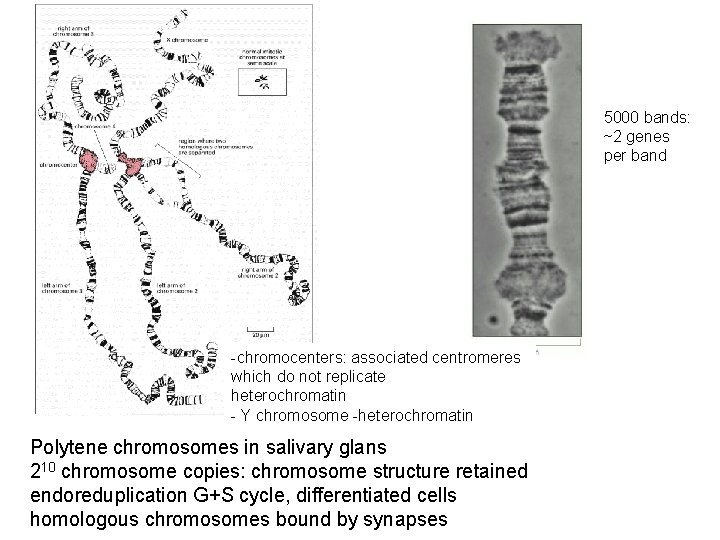
5000 bands: ~2 genes per band -chromocenters: associated centromeres which do not replicate heterochromatin - Y chromosome -heterochromatin Polytene chromosomes in salivary glans 210 chromosome copies: chromosome structure retained endoreduplication G+S cycle, differentiated cells homologous chromosomes bound by synapses
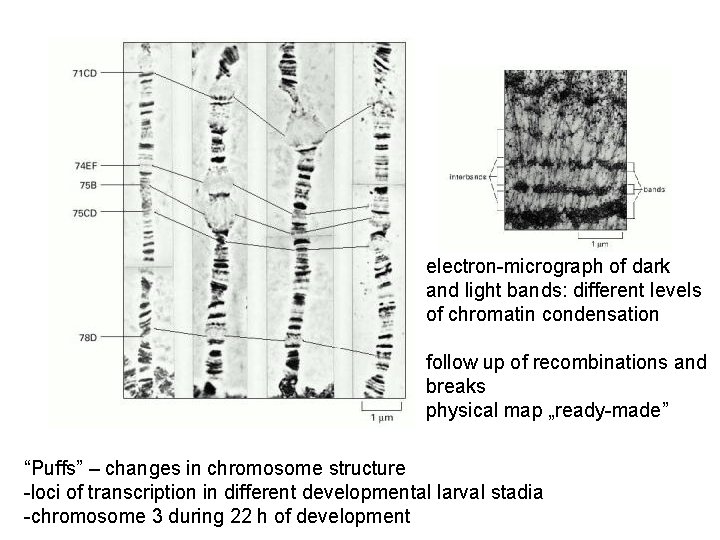
electron-micrograph of dark and light bands: different levels of chromatin condensation follow up of recombinations and breaks physical map „ready-made” “Puffs” – changes in chromosome structure -loci of transcription in different developmental larval stadia -chromosome 3 during 22 h of development
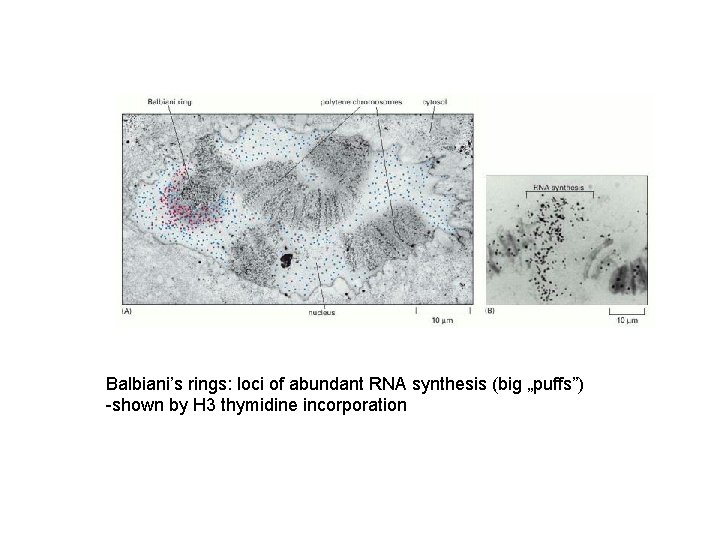
Balbiani’s rings: loci of abundant RNA synthesis (big „puffs”) -shown by H 3 thymidine incorporation
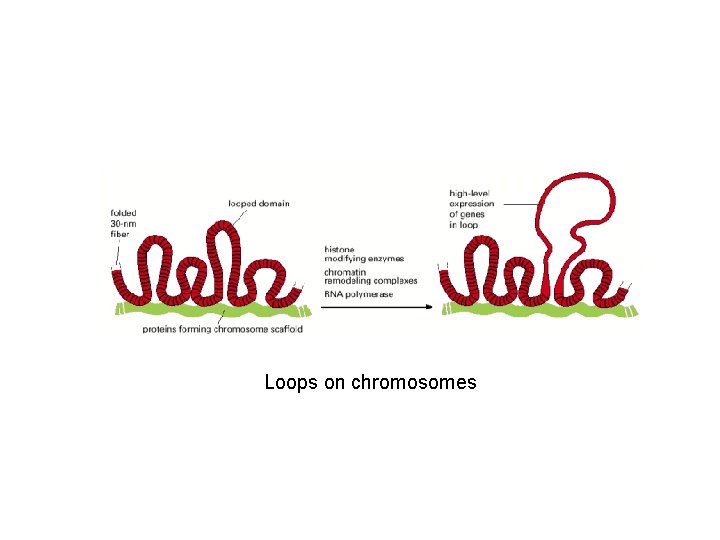
Loops on chromosomes
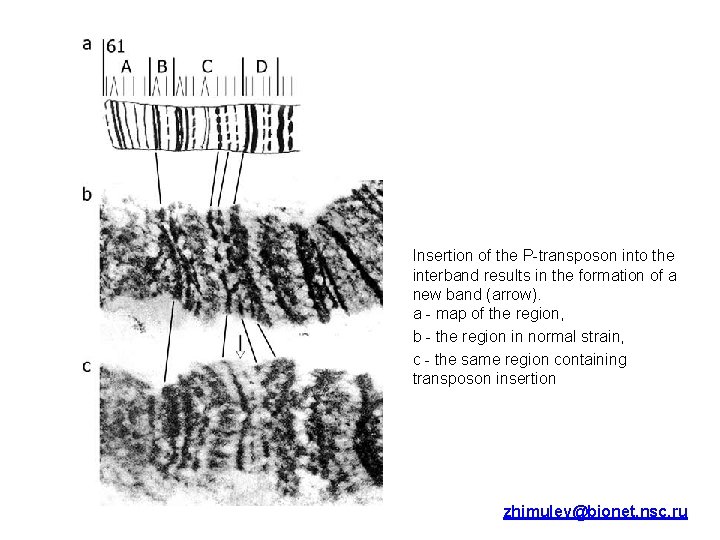
Insertion of the P-transposon into the interband results in the formation of a new band (arrow). a - map of the region, b - the region in normal strain, c - the same region containing transposon insertion zhimulev@bionet. nsc. ru

fruit fly genome 170 000 bp, 13 500 genes (5 % human genome) 21 % repetitive regions, satellite DNA in centromeres and chromosome Y 3 % repetitive r. RNA, 5 S ribosome, histones 9 % 50 families of transposons, in centromeres and telomeres 67 % unique DNA

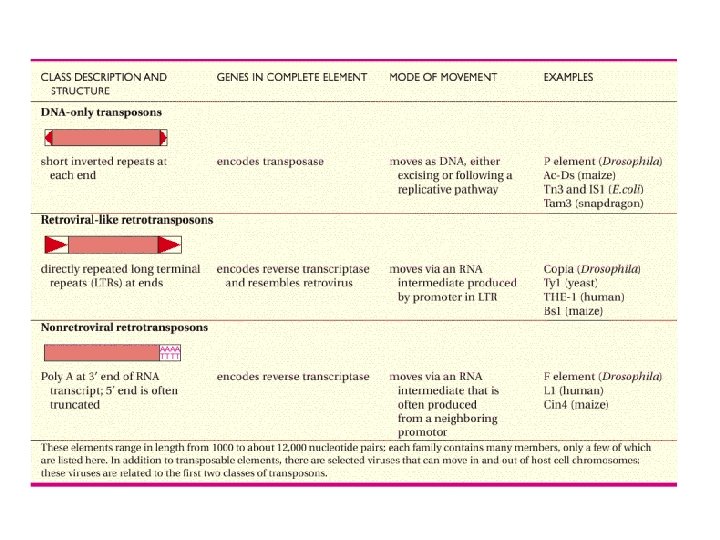
Transposons: movable genetic elements DNA fragments which can change their position in the genome
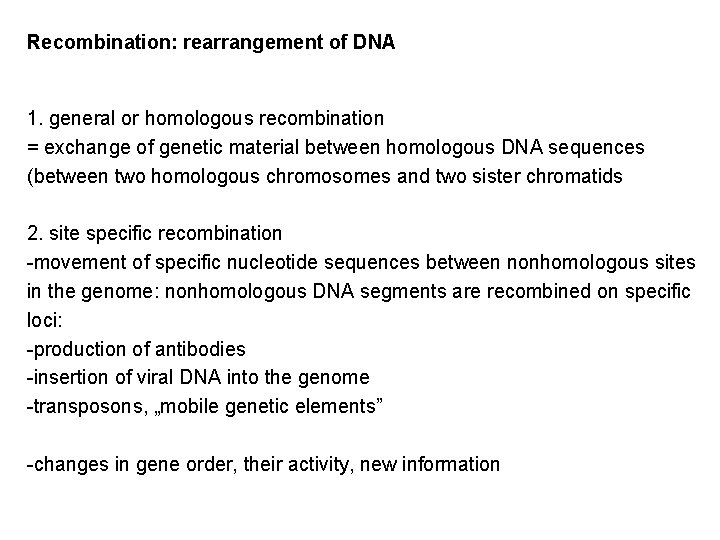
Recombination: rearrangement of DNA 1. general or homologous recombination = exchange of genetic material between homologous DNA sequences (between two homologous chromosomes and two sister chromatids 2. site specific recombination -movement of specific nucleotide sequences between nonhomologous sites in the genome: nonhomologous DNA segments are recombined on specific loci: -production of antibodies -insertion of viral DNA into the genome -transposons, „mobile genetic elements” -changes in gene order, their activity, new information

Mobile genetic elements DNA fragments which can change their loci in the genome -found in the genomes of all the species -from several tens to over 10000 bp -moving inside and between chromosomes -viruses can exit from the cell, transposons not -often in high number of copies (>50% human genome, >80% maize genome), but many are defect -transposition in humans in germ cells: in 1 of 8 persons -translocations can cause deletions and rearrangements; possible role in evolution

Site-specific recombinations -transposition: break at the ends of mobile fragment on the chromosome and on the target site which is not homologous -conservative recombination: small homologous region, short heteroduplex
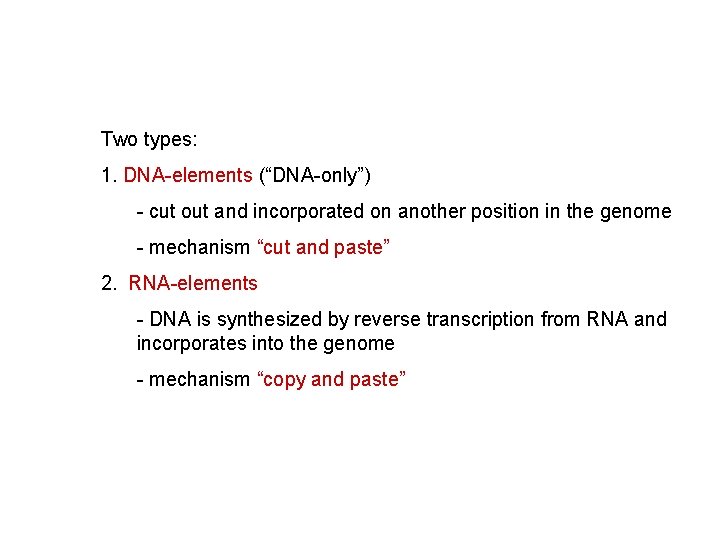
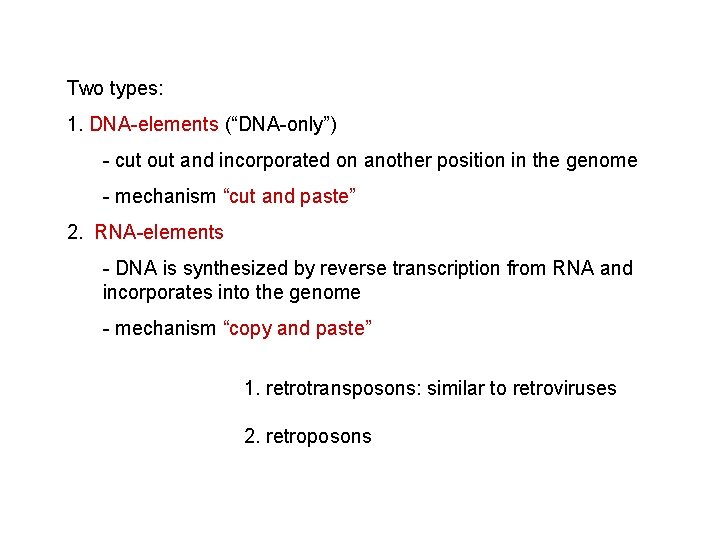
Two types: 1. DNA-elements (“DNA-only”) - cut out and incorporated on another position in the genome - mechanism “cut and paste” 2. RNA-elements - DNA is synthesized by reverse transcription from RNA and incorporates into the genome - mechanism “copy and paste”
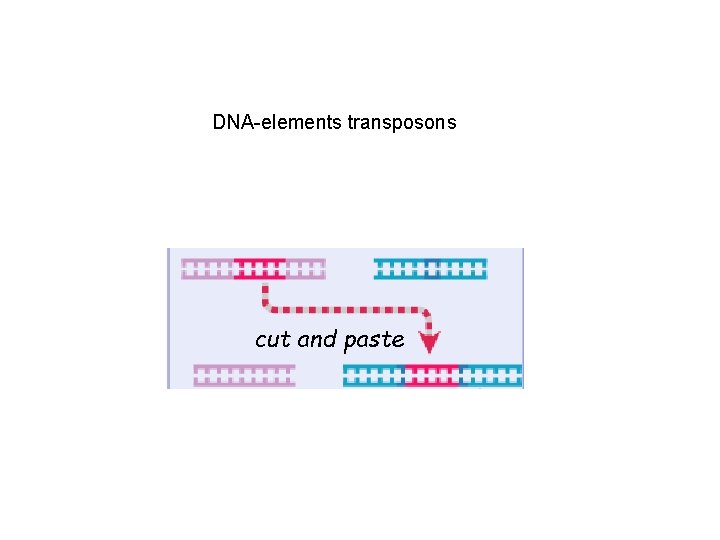
DNA-elements transposons cut and paste
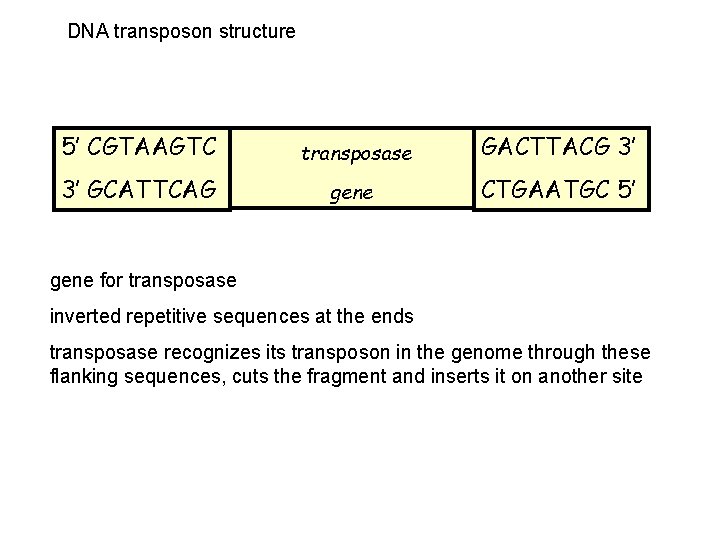
DNA transposon structure 5’ CGTAAGTC transposase GACTTACG 3’ 3’ GCATTCAG gene CTGAATGC 5’ gene for transposase inverted repetitive sequences at the ends transposase recognizes its transposon in the genome through these flanking sequences, cuts the fragment and inserts it on another site
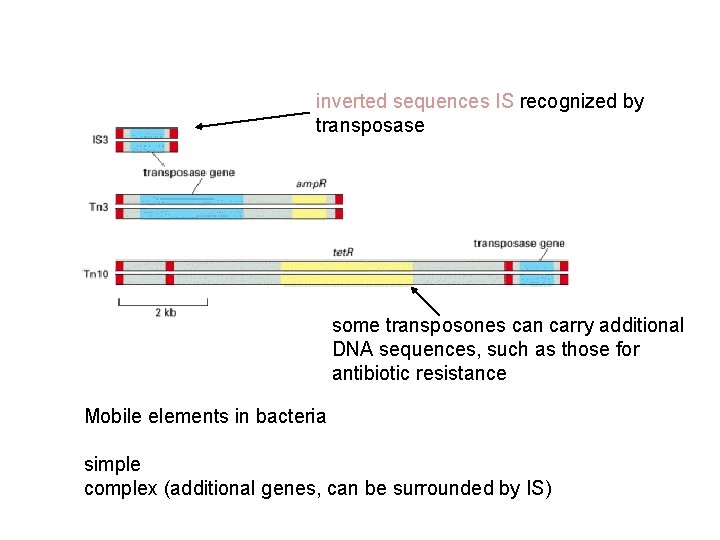
inverted sequences IS recognized by transposase some transposones can carry additional DNA sequences, such as those for antibiotic resistance Mobile elements in bacteria simple complex (additional genes, can be surrounded by IS)
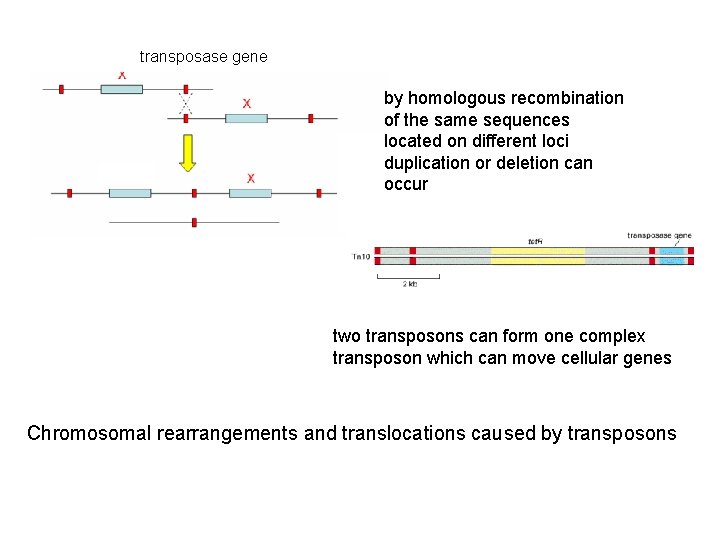
gen transposase gene by homologous recombination of the same sequences located on different loci duplication or deletion can occur two transposons can form one complex transposon which can move cellular genes Chromosomal rearrangements and translocations caused by transposons

DNA damage repair by homologous and nonhomologous recombination Transposition “cut and paste” -IS of 20 nt are recognized by transposase dimer -they form loop and cut out the fragment; chromosome is repaired -on target chromosome transposase does staggered cleavage and inserts the transposon -single stranded flanked ends ( ~5 -8 nt) are filled in by DNA polymerase -direct repetition of the host DNA are left behind after transposition
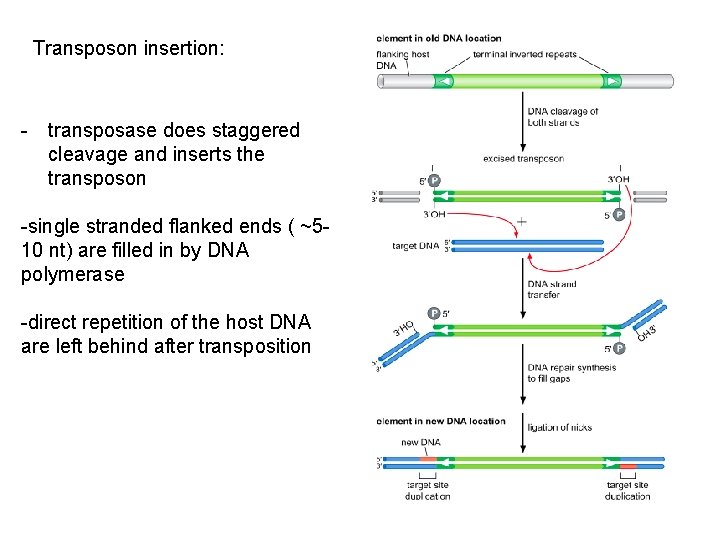
Transposon insertion: - transposase does staggered cleavage and inserts the transposon -single stranded flanked ends ( ~510 nt) are filled in by DNA polymerase -direct repetition of the host DNA are left behind after transposition
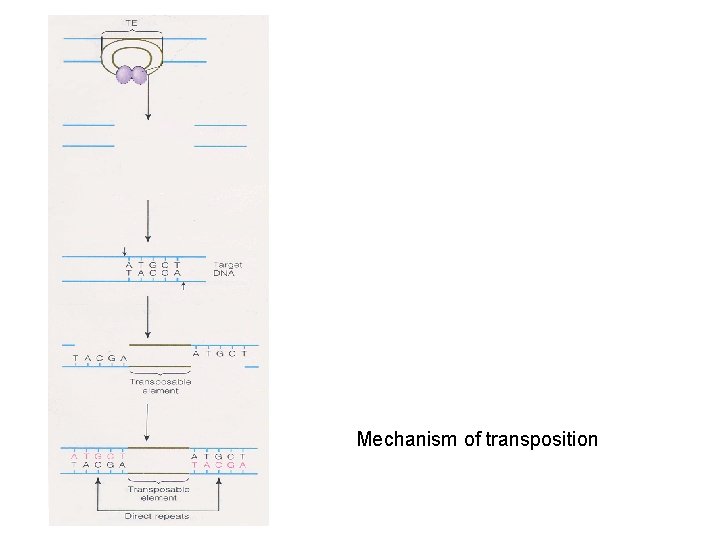
Mechanism of transposition
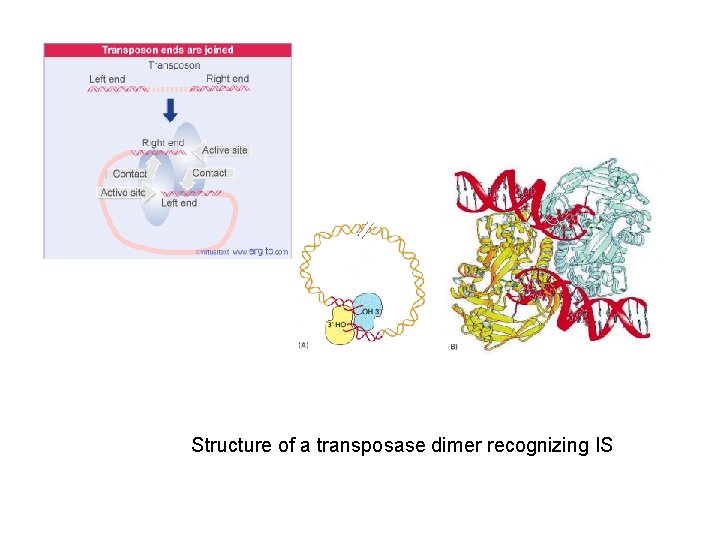
Structure of a transposase dimer recognizing IS
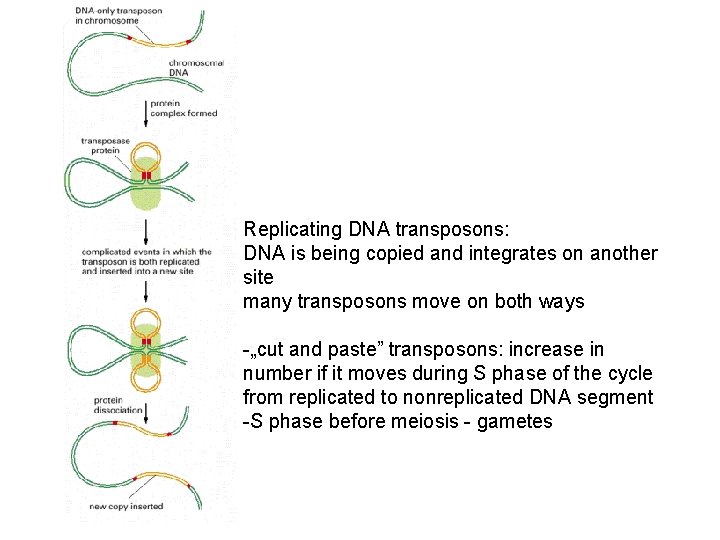
Replicating DNA transposons: DNA is being copied and integrates on another site many transposons move on both ways -„cut and paste” transposons: increase in number if it moves during S phase of the cycle from replicated to nonreplicated DNA segment -S phase before meiosis - gametes
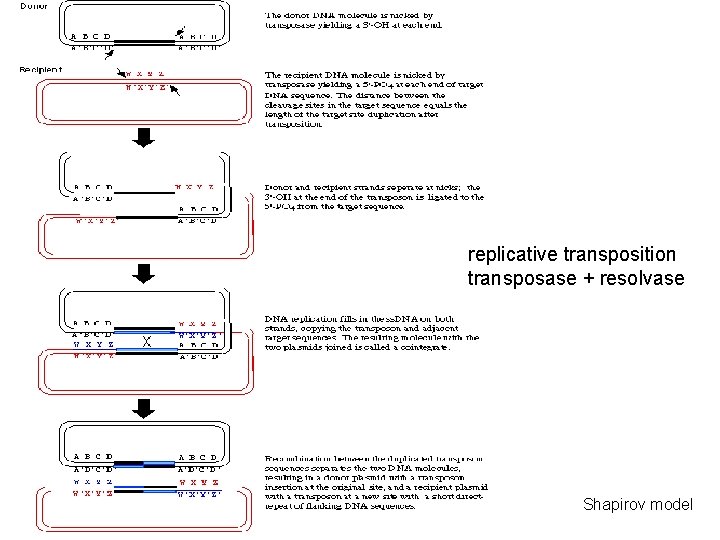
replicative transposition transposase + resolvase Shapirov model
Two types: 1. DNA-elements (“DNA-only”) - cut out and incorporated on another position in the genome - mechanism “cut and paste” 2. RNA-elements - DNA is synthesized by reverse transcription from RNA and incorporates into the genome - mechanism “copy and paste” 1. retrotransposons: similar to retroviruses 2. retroposons
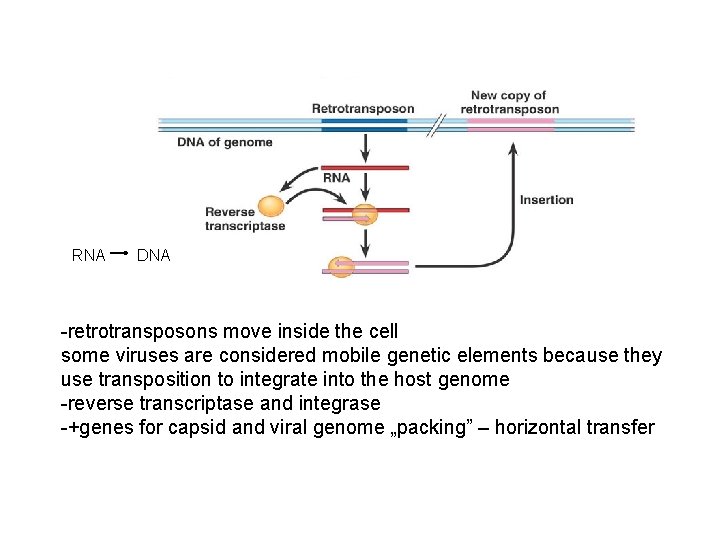
RNA DNA -retrotransposons move inside the cell some viruses are considered mobile genetic elements because they use transposition to integrate into the host genome -reverse transcriptase and integrase -+genes for capsid and viral genome „packing” – horizontal transfer
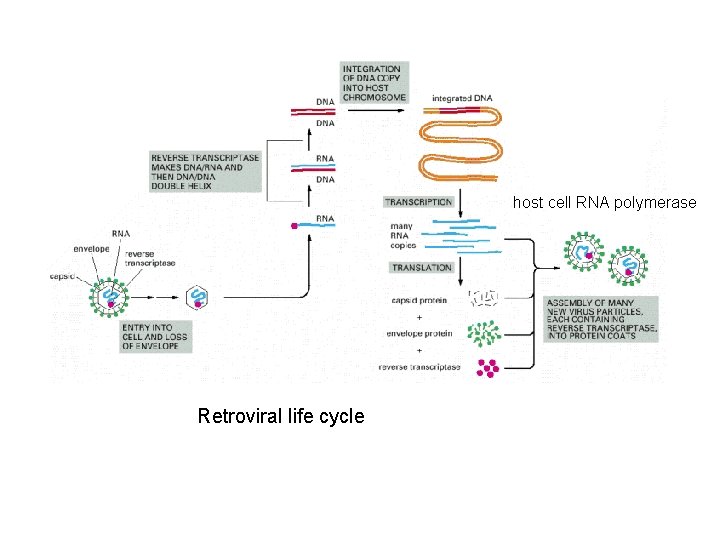
host cell RNA polymerase Retroviral life cycle
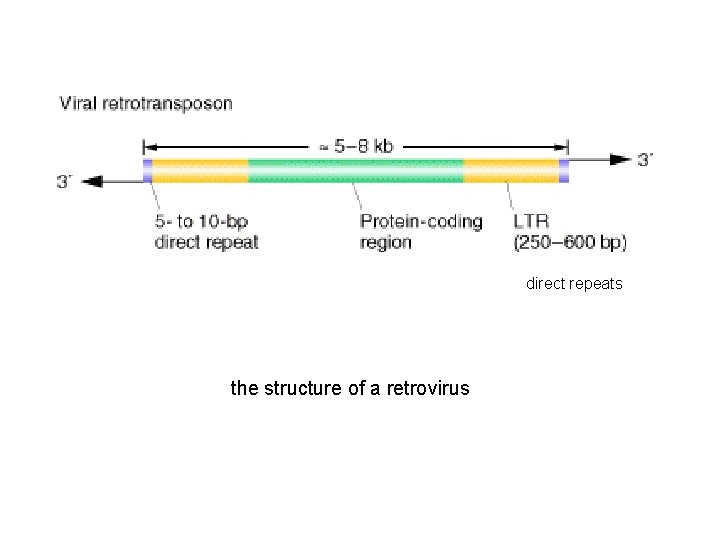
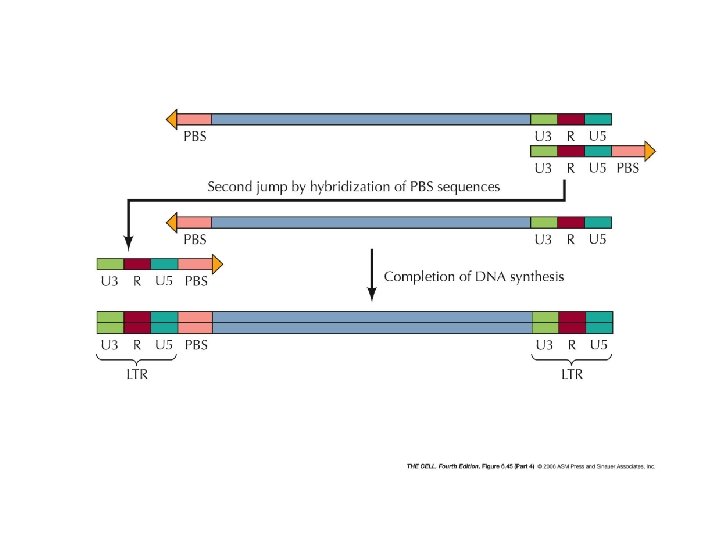
direct repeats the structure of a retrovirus
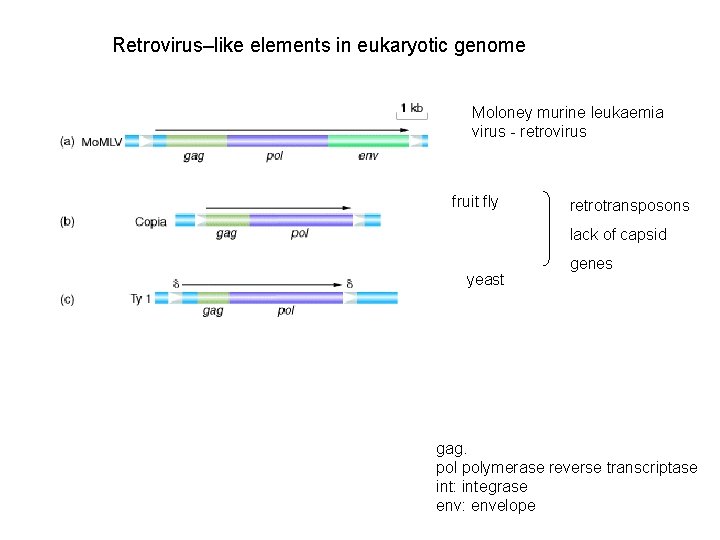
Retrovirus–like elements in eukaryotic genome Moloney murine leukaemia virus - retrovirus fruit fly retrotransposons lack of capsid yeast genes humani LINE ga gag. polymerase reverse transcriptase int: integrase env: envelope
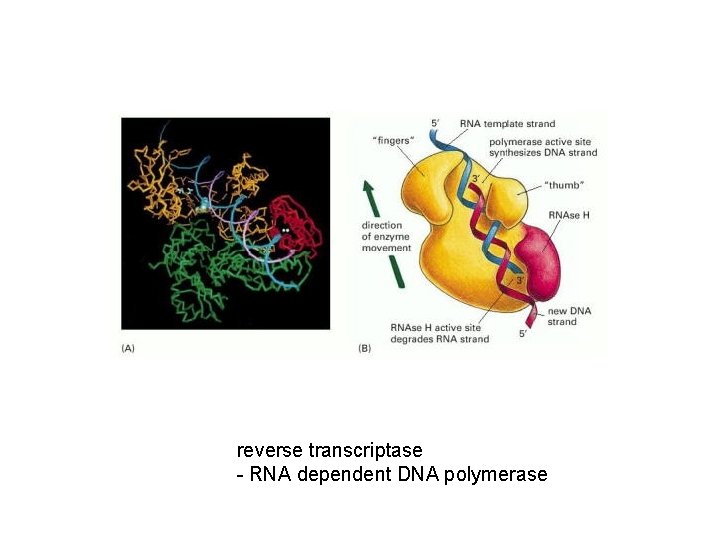
reverse transcriptase - RNA dependent DNA polymerase
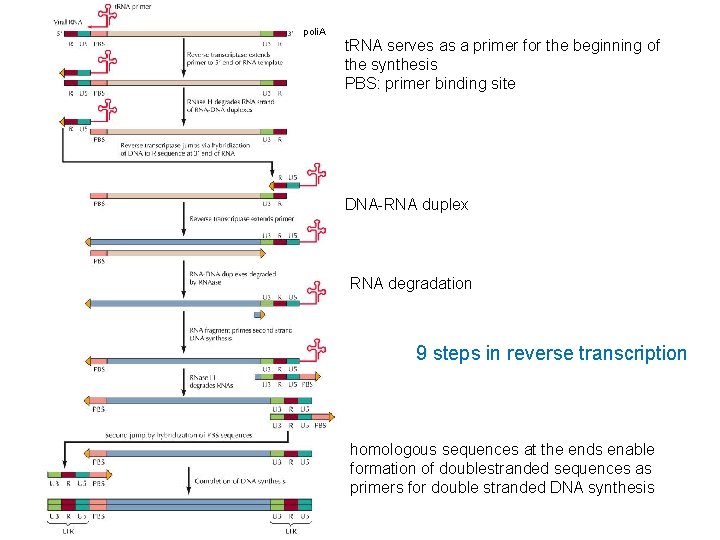
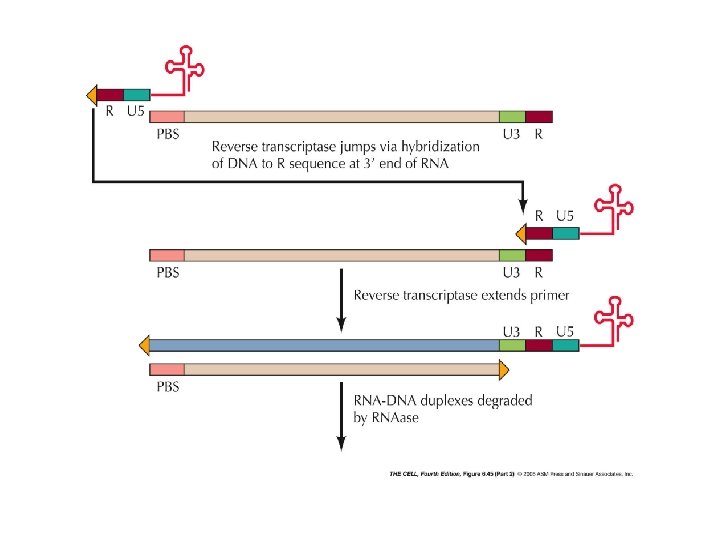
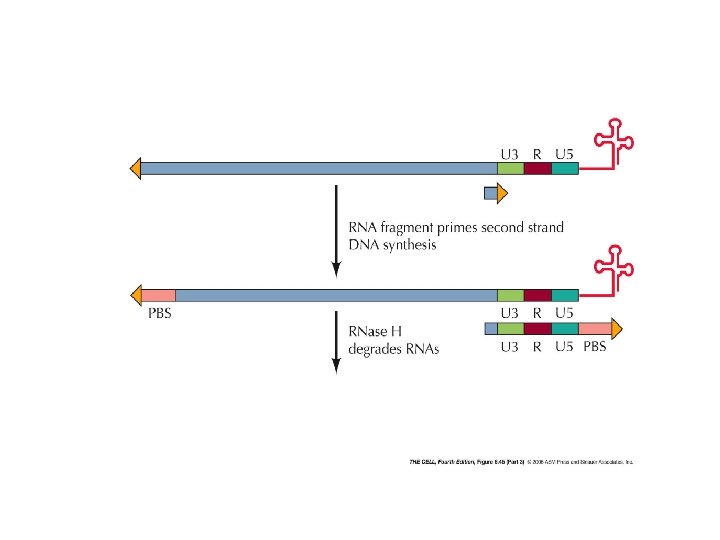
poli. A t. RNA serves as a primer for the beginning of the synthesis PBS: primer binding site DNA-RNA duplex RNA degradation 9 steps in reverse transcription homologous sequences at the ends enable formation of doublestranded sequences as primers for double stranded DNA synthesis
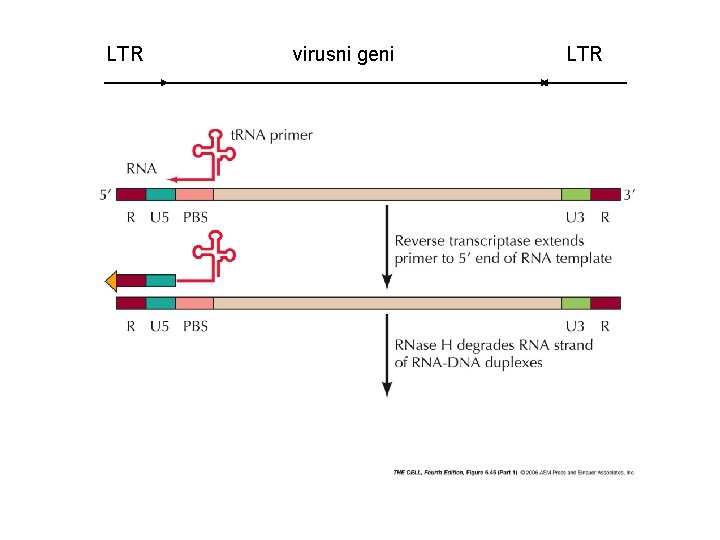
LTR virusni geni LTR
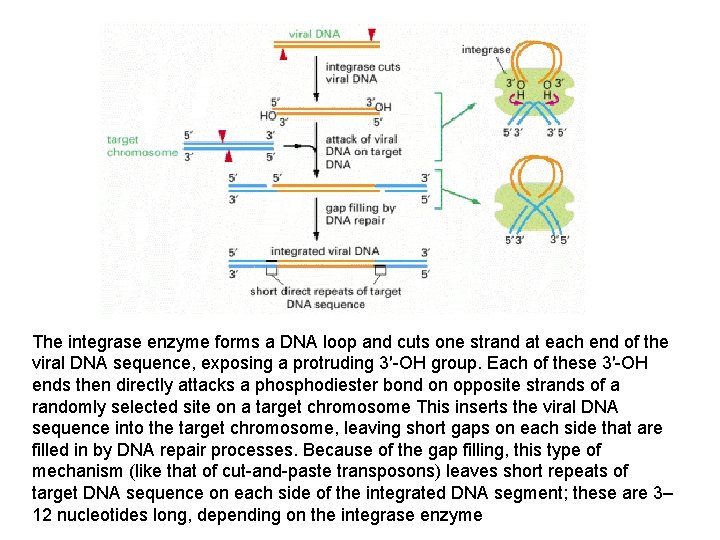
The integrase enzyme forms a DNA loop and cuts one strand at each end of the viral DNA sequence, exposing a protruding 3′-OH group. Each of these 3′-OH ends then directly attacks a phosphodiester bond on opposite strands of a randomly selected site on a target chromosome This inserts the viral DNA sequence into the target chromosome, leaving short gaps on each side that are filled in by DNA repair processes. Because of the gap filling, this type of mechanism (like that of cut-and-paste transposons) leaves short repeats of target DNA sequence on each side of the integrated DNA segment; these are 3– 12 nucleotides long, depending on the integrase enzyme
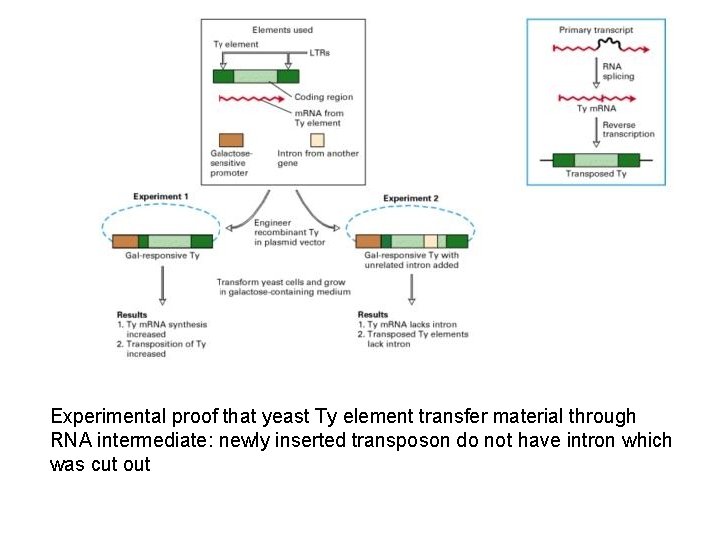
Experimental proof that yeast Ty element transfer material through RNA intermediate: newly inserted transposon do not have intron which was cut out

Retrotransposons 1. LTR retrotransposons 2. retroposons: -LINE (long interspersed elements) interspersed repetitive nucleotide sequences longer then 5000 bp present in more then 104 copies in the genome. SINE (short interspersed elements) interspersed repetitive nucleotide sequences shorter then 500 bp, present in more then 105 copies in the genome.
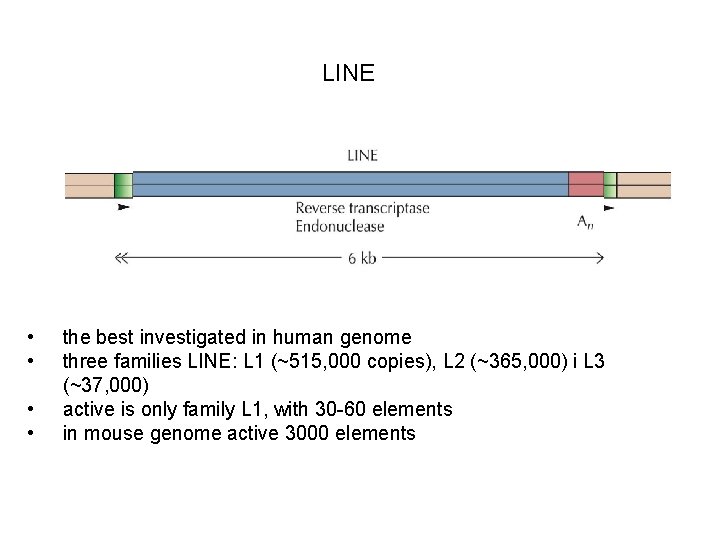
LINE • • the best investigated in human genome three families LINE: L 1 (~515, 000 copies), L 2 (~365, 000) i L 3 (~37, 000) active is only family L 1, with 30 -60 elements in mouse genome active 3000 elements
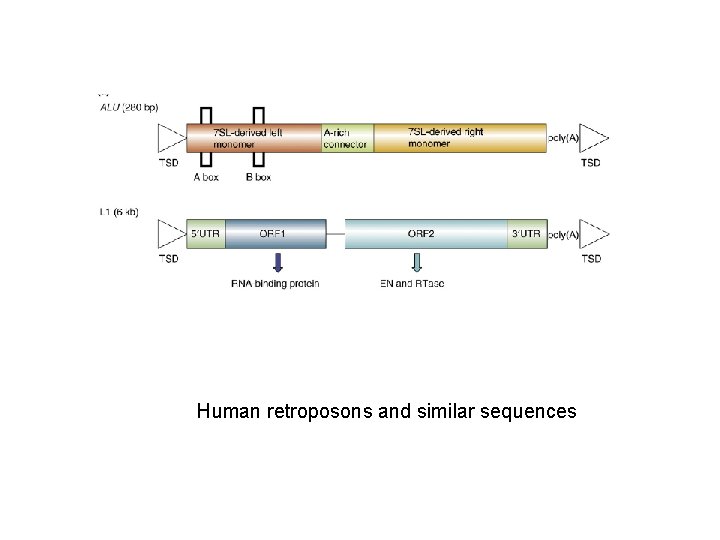
Human retroposons and similar sequences
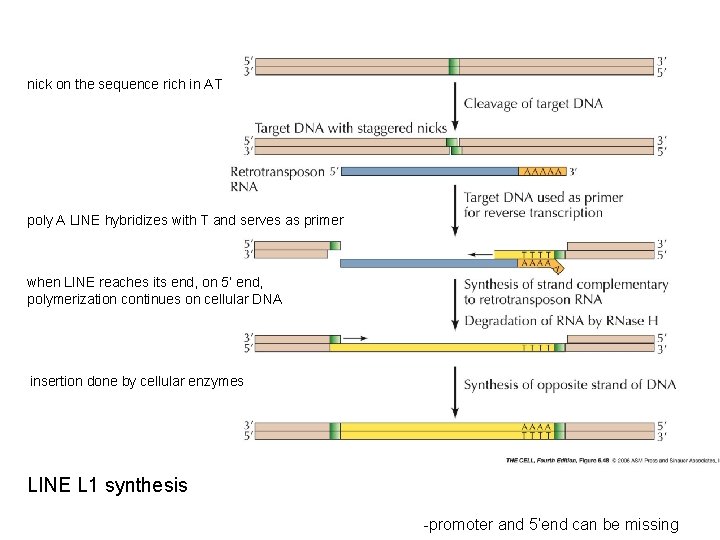
nick on the sequence rich in AT poly A LINE hybridizes with T and serves as primer when LINE reaches its end, on 5’ end, polymerization continues on cellular DNA insertion done by cellular enzymes LINE L 1 synthesis -promoter and 5’end can be missing
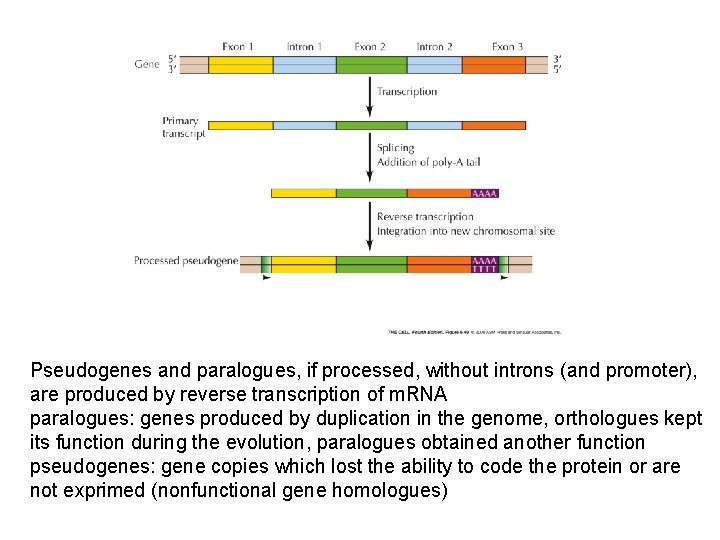
Pseudogenes and paralogues, if processed, without introns (and promoter), are produced by reverse transcription of m. RNA paralogues: genes produced by duplication in the genome, orthologues kept its function during the evolution, paralogues obtained another function pseudogenes: gene copies which lost the ability to code the protein or are not exprimed (nonfunctional gene homologues)
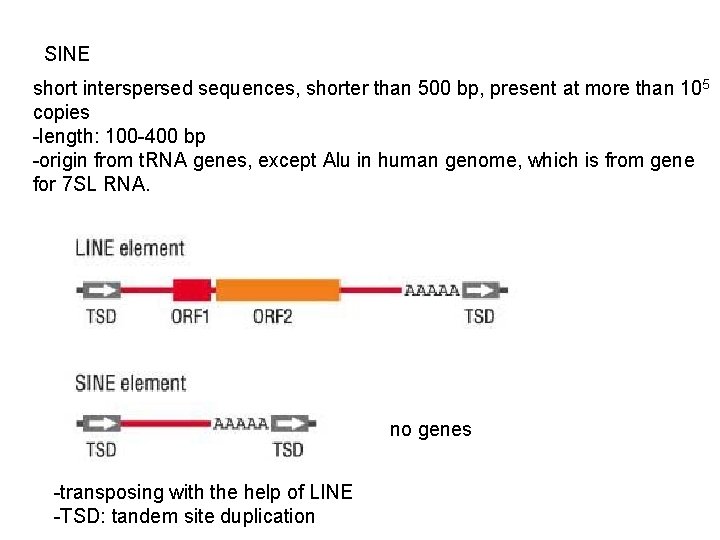
SINE short interspersed sequences, shorter than 500 bp, present at more than 105 copies -length: 100 -400 bp -origin from t. RNA genes, except Alu in human genome, which is from gene for 7 SL RNA. no genes -transposing with the help of LINE -TSD: tandem site duplication
-transposition in bacteria is rare: 1 in 105 generations -depends on specific gene expression in the cell -environmental factors (stress, temperature, pathogens, mutagens) -interactions between cellular and environmental factors -expansion in certain phases of species evolution -after that period, expansion stops and optimal number of transposons is established, for the species and the conditions -random insertion site, active transposons are NOT in heterochromatin -sites of integration are usually rich in CG

-great part of the genome in vertebrates is made of repetitive sequences -in human chromosomes there are mutated and deleted sequences of retroposon L 1 they are called LINE L 1 are mostly nonmobile, but some can move translocation of L 1 into the gene for coagulation factor VIII hemophilia -L 1 can move by itself, shorter sequences can use its enzyme
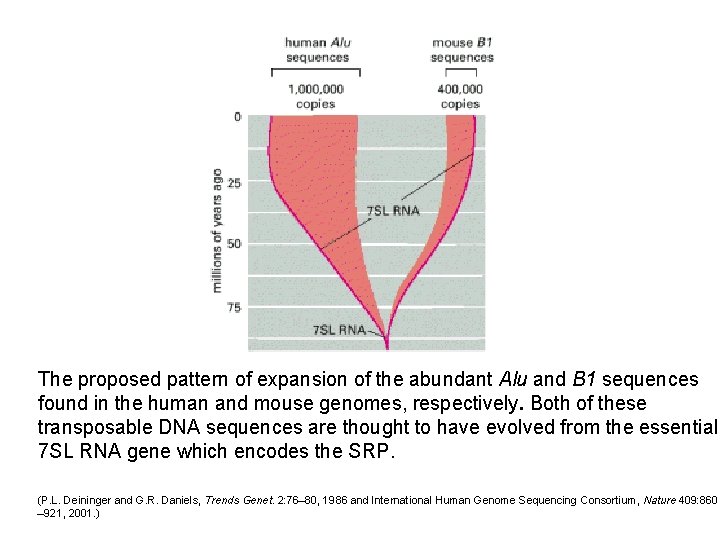
The proposed pattern of expansion of the abundant Alu and B 1 sequences found in the human and mouse genomes, respectively. Both of these transposable DNA sequences are thought to have evolved from the essential 7 SL RNA gene which encodes the SRP. (P. L. Deininger and G. R. Daniels, Trends Genet. 2: 76– 80, 1986 and International Human Genome Sequencing Consortium, Nature 409: 860 – 921, 2001. )

-in different organisms, different types of mobile elements dominate: bacteria: DNA transposons yeast: retroviral retrotransposons human: all three types -DNA-only transposons: active at the time before divergence between humans and monkeys, today they accumulated mutations and are not active retroviral retrotransposons: active only one family, others inactivated by mutations nonretroviral retrotransposons: only active and can cause mutations, 2 in 1000 -role in evolution: activity at the time of speciation (hypothesis) „selfish DNA exon shuffling: insertion of exons in genes – new proteins changes in expression, insertions, changes in promoters, inactivation
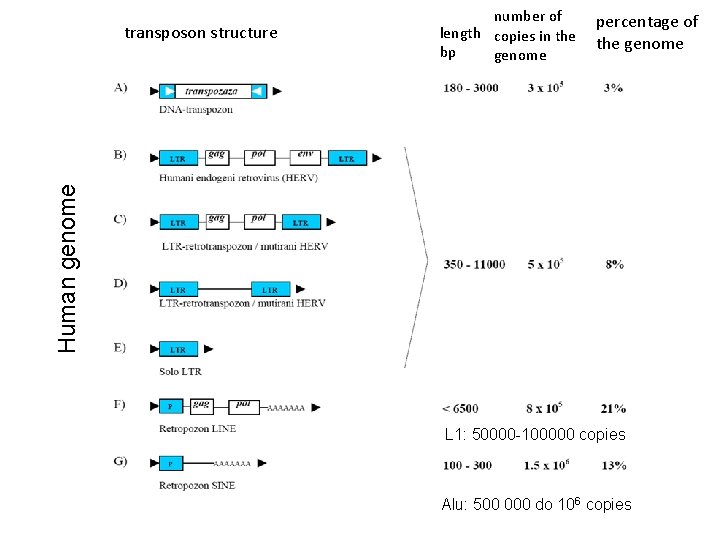
percentage of the genome Human genome transposon structure number of length copies in the bp genome L 1: 50000 -100000 copies Alu: 500 000 do 106 copies

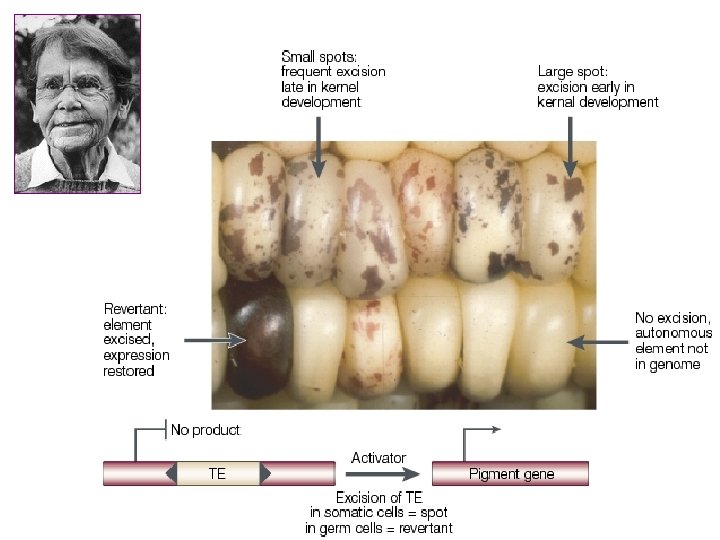
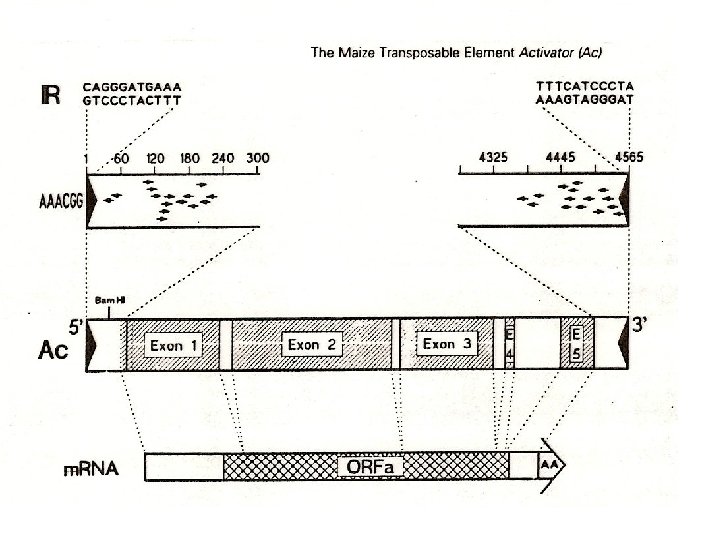
Barbara Mc. Clintock (1902 -1992) discovered transposons (jumping genes) in maise in 1940. She called them activator (Ac) Ac and dissociation (Ds) Ds elements. For twenty years nobody took her experiments for serious but she got Nobel prize in 1983
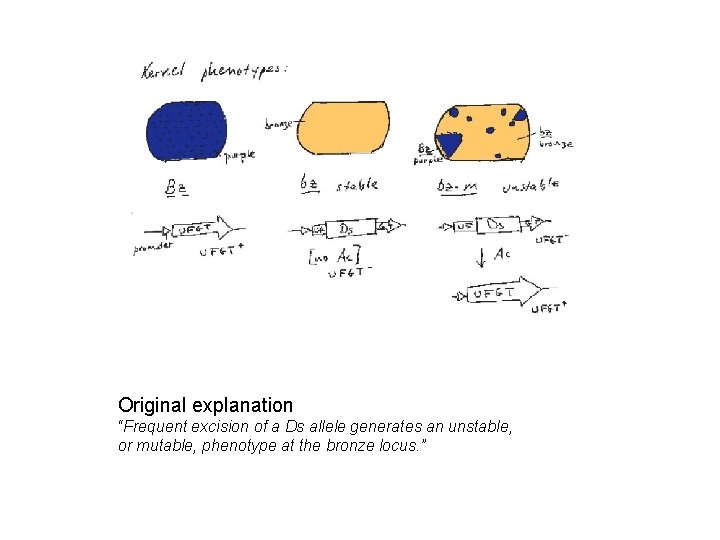
Original explanation “Frequent excision of a Ds allele generates an unstable, or mutable, phenotype at the bronze locus. ”
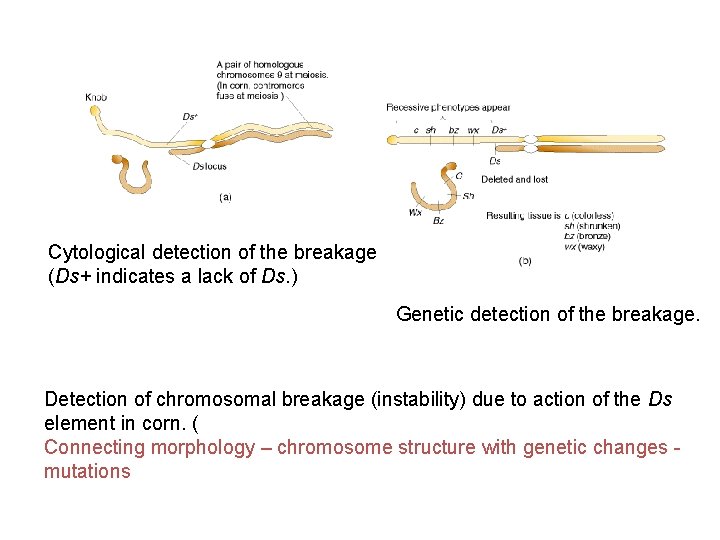
Cytological detection of the breakage (Ds+ indicates a lack of Ds. ) Genetic detection of the breakage. Detection of chromosomal breakage (instability) due to action of the Ds element in corn. ( Connecting morphology – chromosome structure with genetic changes mutations
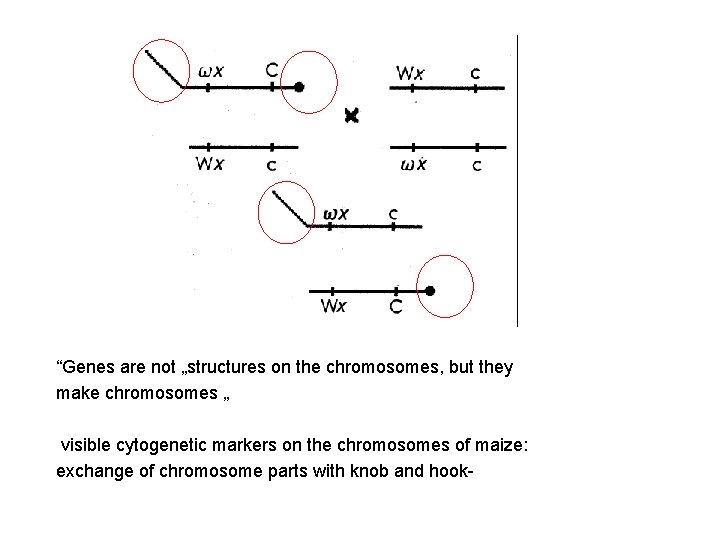
“Genes are not „structures on the chromosomes, but they make chromosomes „ visible cytogenetic markers on the chromosomes of maize: exchange of chromosome parts with knob and hook-
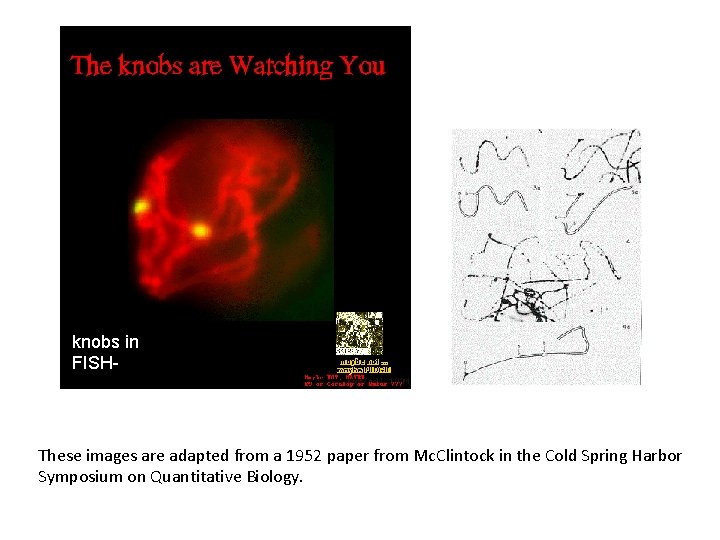
knobs in FISH- These images are adapted from a 1952 paper from Mc. Clintock in the Cold Spring Harbor Symposium on Quantitative Biology.
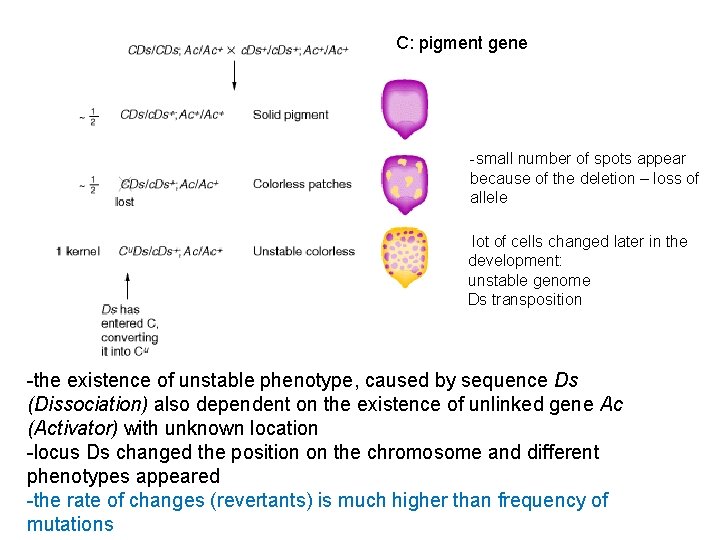
C: pigment gene -small number of spots appear because of the deletion – loss of allele lot of cells changed later in the development: unstable genome Ds transposition -the existence of unstable phenotype, caused by sequence Ds (Dissociation) also dependent on the existence of unlinked gene Ac (Activator) with unknown location -locus Ds changed the position on the chromosome and different phenotypes appeared -the rate of changes (revertants) is much higher than frequency of mutations
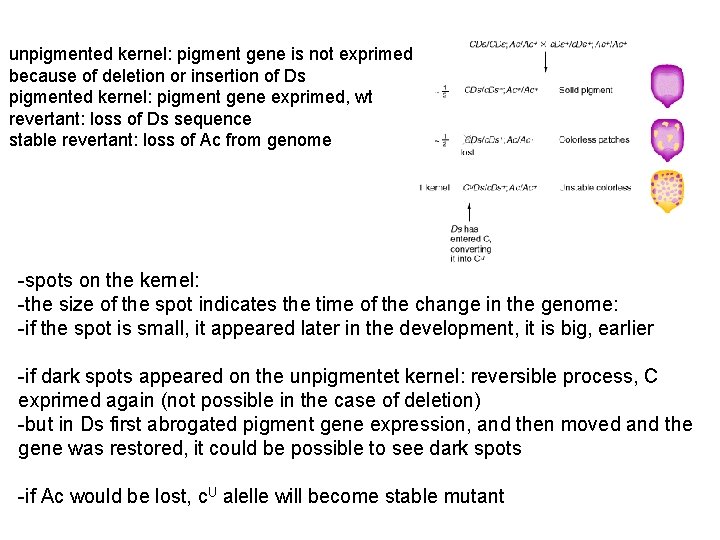
unpigmented kernel: pigment gene is not exprimed because of deletion or insertion of Ds pigmented kernel: pigment gene exprimed, wt revertant: loss of Ds sequence stable revertant: loss of Ac from genome -spots on the kernel: -the size of the spot indicates the time of the change in the genome: -if the spot is small, it appeared later in the development, it is big, earlier -if dark spots appeared on the unpigmentet kernel: reversible process, C exprimed again (not possible in the case of deletion) -but in Ds first abrogated pigment gene expression, and then moved and the gene was restored, it could be possible to see dark spots -if Ac would be lost, c. U alelle will become stable mutant
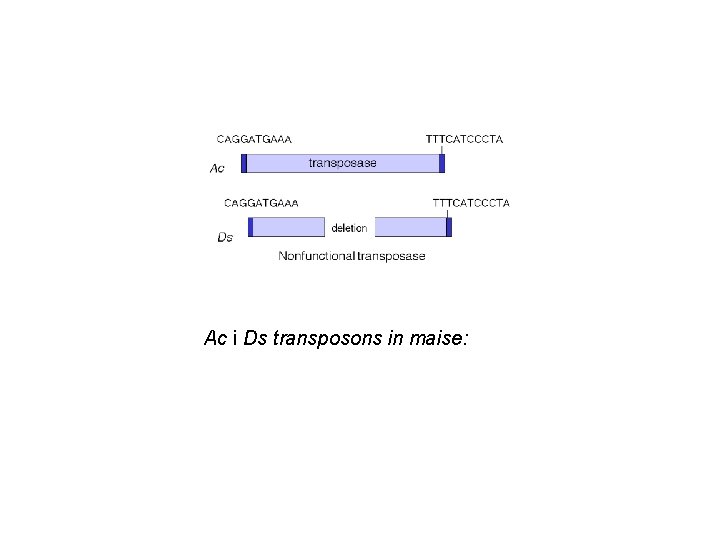
Ac i Ds transposons in maise:
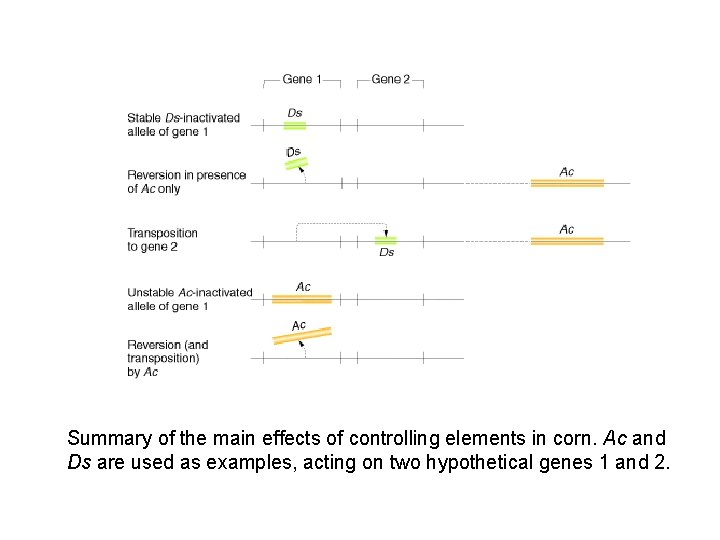
Summary of the main effects of controlling elements in corn. Ac and Ds are used as examples, acting on two hypothetical genes 1 and 2.
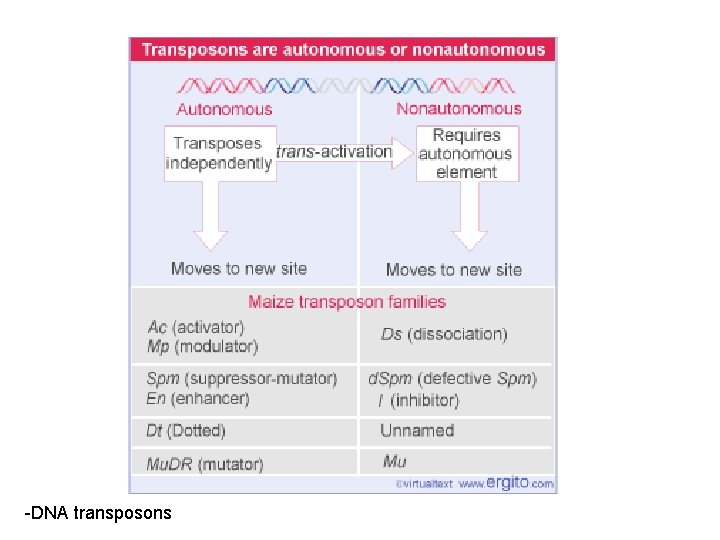
-DNA transposons

Fruit fly and P element

D. melanogaster is now a cosmopolitan species, but it is thought to have evolved in western Africa. The species became established elsewhere only when human commercial shipping provided a means for long distance migration. Meanwhile, D. willistoni and related species evolved primarily in Central and South America, and are still endemic to these regions. Therefore, they had no contact with melanogaster until the latter species arrived in the Americas. Johnson (1913), by examining antique insect collections, estimates that the first appearance of D. melanogaster in the New World occurred in the early 1800's, and they became widespread by the end of the century. The horizontal transfer event could have occurred at any time since melanogaster and willistoni became sympatric, but the spread of P elements through melanogaster was presumably not yet complete by the 1930's when the last laboratory M populations were established.
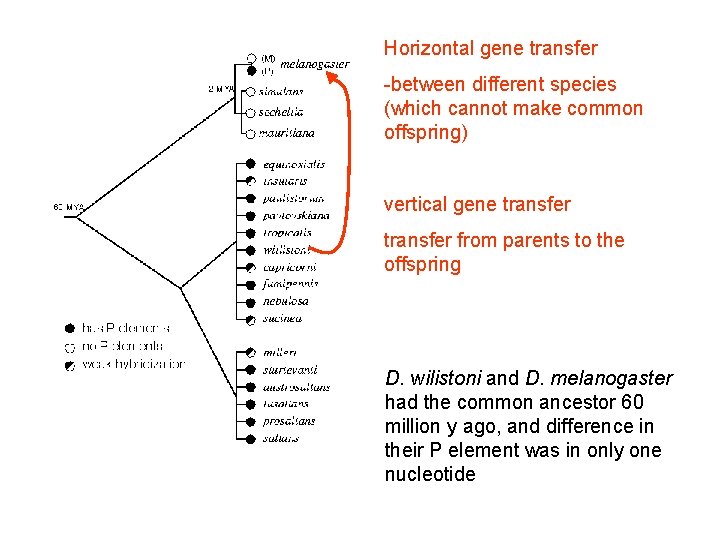
Horizontal gene transfer -between different species (which cannot make common offspring) vertical gene transfer from parents to the offspring D. wilistoni and D. melanogaster had the common ancestor 60 million y ago, and difference in their P element was in only one nucleotide
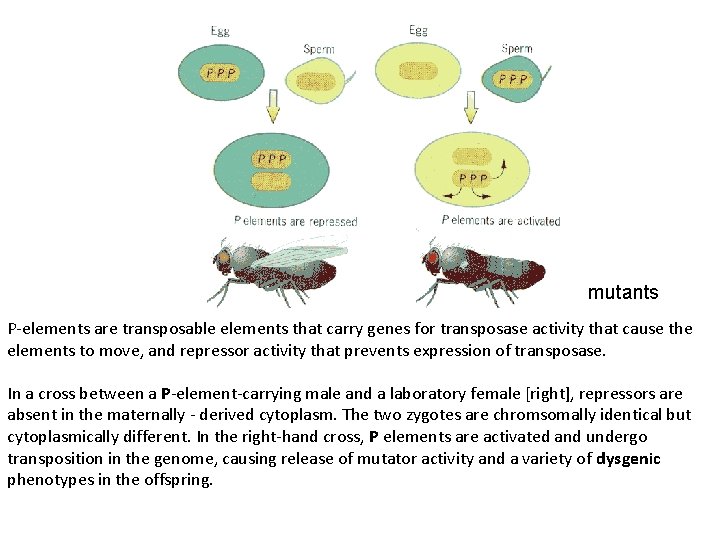
mutants P-elements are transposable elements that carry genes for transposase activity that cause the elements to move, and repressor activity that prevents expression of transposase. In a cross between a P-element-carrying male and a laboratory female [right], repressors are absent in the maternally - derived cytoplasm. The two zygotes are chromsomally identical but cytoplasmically different. In the right-hand cross, P elements are activated and undergo transposition in the genome, causing release of mutator activity and a variety of dysgenic phenotypes in the offspring.
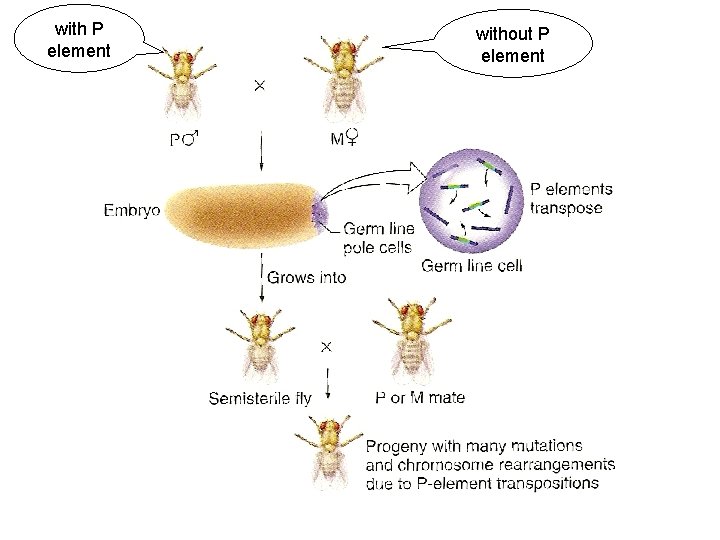
with P element without P element
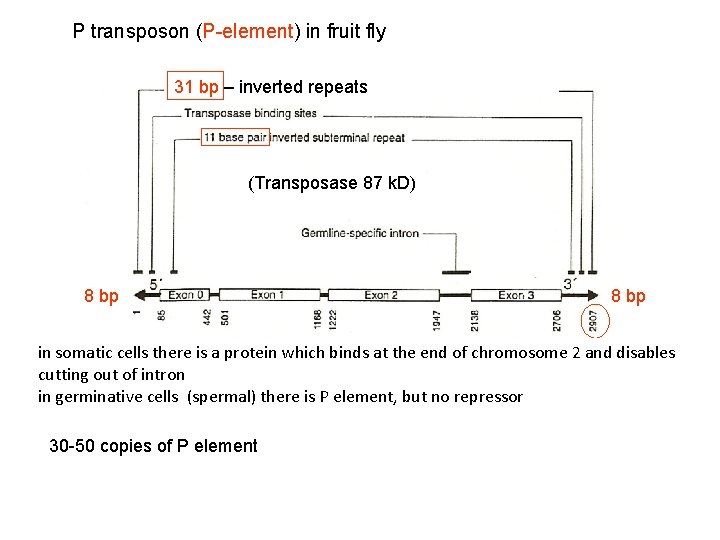
P transposon (P-element) in fruit fly 31 bp – inverted repeats (Transposase 87 k. D) 8 bp in somatic cells there is a protein which binds at the end of chromosome 2 and disables cutting out of intron in germinative cells (spermal) there is P element, but no repressor 30 -50 copies of P element
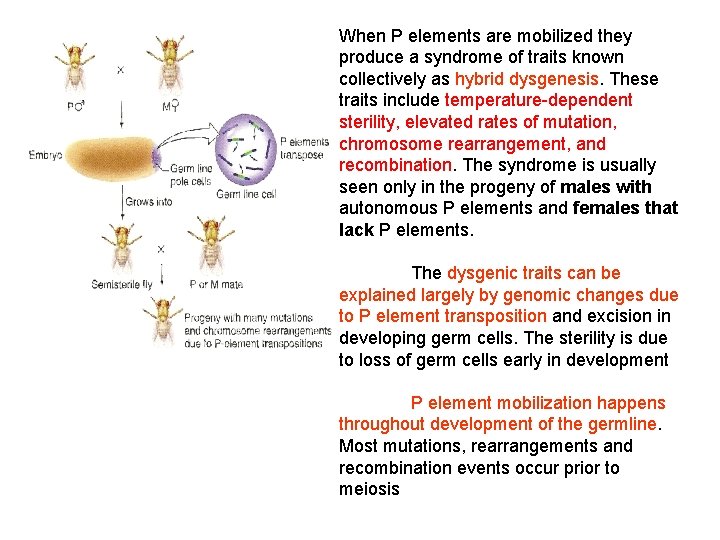
When P elements are mobilized they produce a syndrome of traits known collectively as hybrid dysgenesis. These traits include temperature-dependent sterility, elevated rates of mutation, chromosome rearrangement, and recombination. The syndrome is usually seen only in the progeny of males with autonomous P elements and females that lack P elements. The dysgenic traits can be explained largely by genomic changes due to P element transposition and excision in developing germ cells. The sterility is due to loss of germ cells early in development P element mobilization happens throughout development of the germline. Most mutations, rearrangements and recombination events occur prior to meiosis
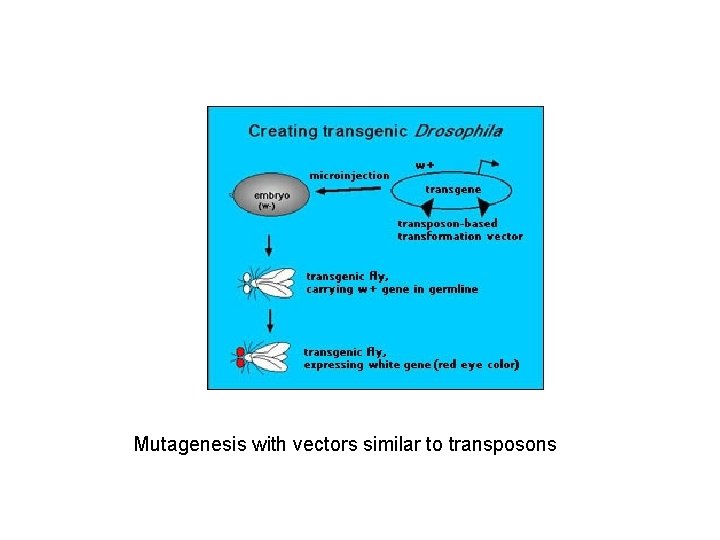
Mutagenesis with vectors similar to transposons
Learning to Fly: Phenotypic Markers in Drosophila. " A poster of common phenotypic markers used in Drosophila genetics. Jennifer Childress, Richard Behringer, and Georg Halder. 2005. Genesis 43(1). Cover illustration.

Image: Scanning electron micrograph of the Drosophila melanogaster sestrin-null mutant used to study pathways involved in oxidative stress and aging. Living and uncoated specimen imaged by SEM Time Flies By Thomas Deerinck, NCMIR

www. hhmi. org/. . . /drosophila-molecular-clock-mode http: //www. hhmi. org/biointeractive/measuring-circadian-activity-drosophila http: //www. dnalc. org/resources/animations/alu. html http: //www. dnalc. org/resources/animations/model_organisms. html https: //www. youtube. com/watch? v=XYZHMGUGq 6 o https: //www. youtube. com/watch? v=_c. Jfs. WYR 42 M&list=PLSp. Pci. Sa. K 5 Ip. R 4 MO 3 r 2 n. RYbz. Sova 5 b. ZRI B. Mc. Clintock https: //www. youtube. com/watch? v=LOGe. Tdcnq. FM – model https: //www. youtube. com/watch? v=Jj 5 Ql. Yl. E 66 w https: //www. youtube. com/watch? v=Crv. K 0 d. Lh. Chw
- Slides: 83