From Protein Sequence to Function Functional Analysis of





![Whole protein functional annotation [Ni. Fe]-hydrogenase maturation factor, carbamoyl phosphate-converting enzyme Acylphosphatase – Znf Whole protein functional annotation [Ni. Fe]-hydrogenase maturation factor, carbamoyl phosphate-converting enzyme Acylphosphatase – Znf](https://slidetodoc.com/presentation_image_h2/19561593889fc34fbf9f79bcece9e9a2/image-47.jpg)






- Slides: 74

From Protein Sequence to Function: Functional Analysis of Protein Sequences and Protein Classification Anastasia Nikolskaya Assistant Professor (Research) Protein Information Resource Department of Biochemistry and Molecular Biology Georgetown University Medical Center

Overview • Role of Bioinformatics/Computational Biology in Proteomics Research • Genomics • Functional Annotation of Proteins • Classification of Proteins Bioinformatics Databases and Analytical Tools: Dr. Mazumder and Dr. Hu Sequence function

Functional Genomics and Proteomics studies biological systems based on global knowledge of protein sets (proteomes). Functional genomics studies biological functions of proteins, complexes, pathways based on the analysis of genome sequences. Includes functional assignments for protein sequences. Genome Transcriptome Proteome Metabolome

Proteomics • Data: Gene Expression Profiling - Genome-Wide Analyses of Gene Expression • Data: Structural Genomics - Determine 3 D Structures of All Protein Families • Data: Genome Projects (Sequencing) - Functional genomics - Knowing complete genome sequences of a number of organisms is the basis of the proteomics research

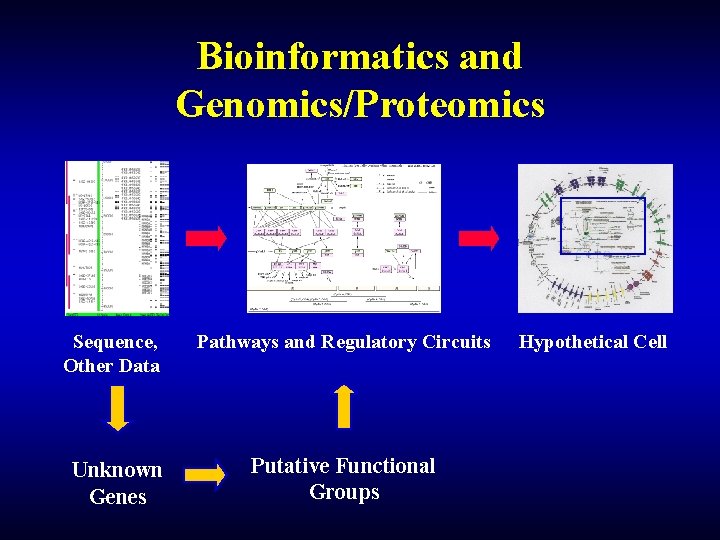
Bioinformatics and Genomics/Proteomics Sequence, Other Data Unknown Genes Pathways and Regulatory Circuits Putative Functional Groups Hypothetical Cell

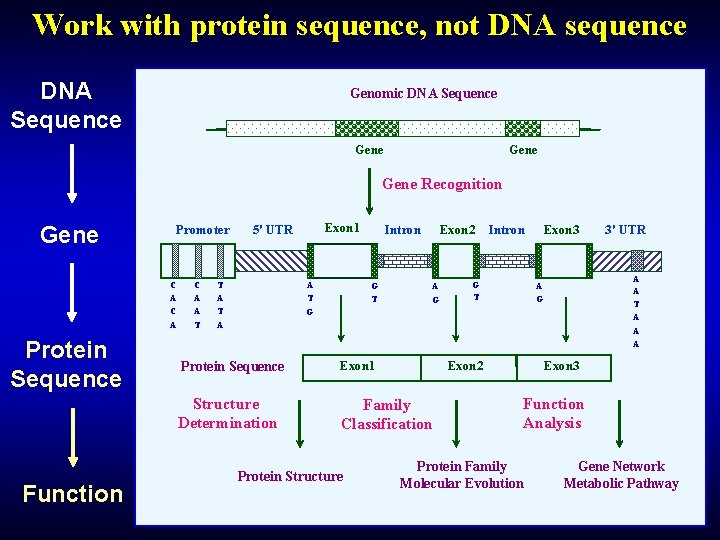
Work with protein sequence, not DNA sequence DNA Sequence Genomic DNA Sequence Gene Recognition Gene Promoter C A Protein Sequence Function C A A T Exon 1 5' UTR T A A T Intron G T Exon 2 A G Exon 3 Intron G T 3' UTR A A T A A A G G A Protein Sequence Exon 1 Structure Determination Family Classification Protein Structure Exon 2 Exon 3 Function Analysis Protein Family Molecular Evolution Gene Network Metabolic Pathway

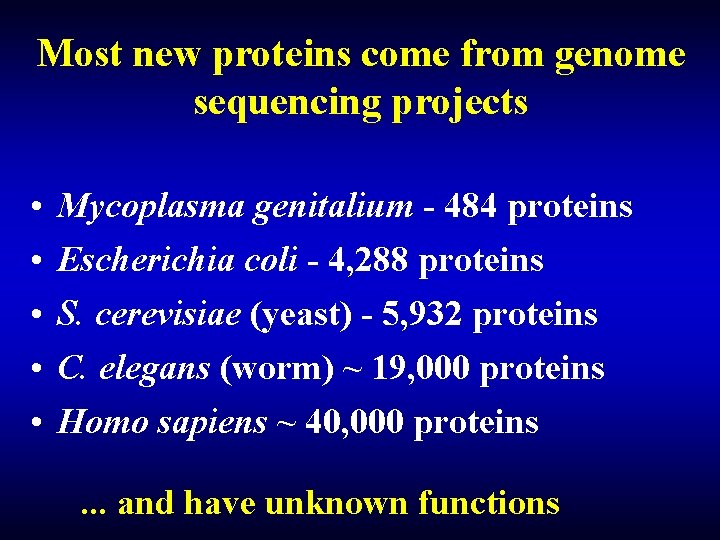
Most new proteins come from genome sequencing projects • • • Mycoplasma genitalium - 484 proteins Escherichia coli - 4, 288 proteins S. cerevisiae (yeast) - 5, 932 proteins C. elegans (worm) ~ 19, 000 proteins Homo sapiens ~ 40, 000 proteins. . . and have unknown functions

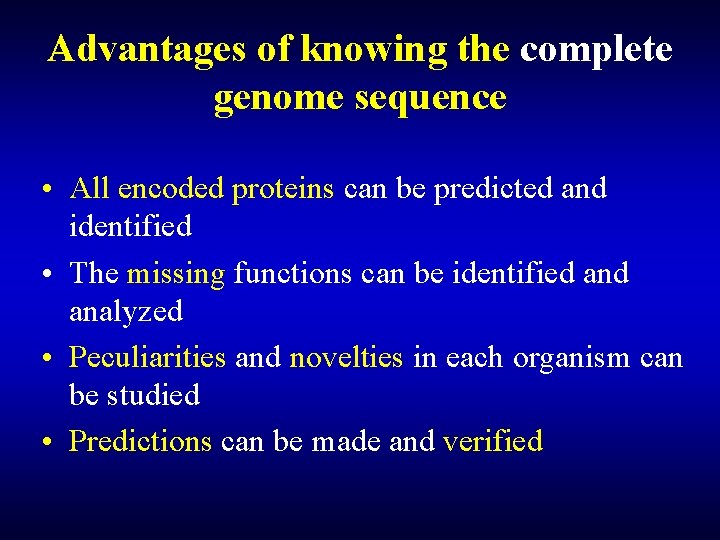
Advantages of knowing the complete genome sequence • All encoded proteins can be predicted and identified • The missing functions can be identified analyzed • Peculiarities and novelties in each organism can be studied • Predictions can be made and verified

The changing face of protein science 20 th century 21 st century • Few well-studied • Many “hypothetiproteins cal” proteins • Mostly globular • Various, often with enzymatic no enzymatic activity • Biased protein • Natural protein set

Properties of the natural protein set • Unexpected diversity of even common enzymes (analogous, paralogous, xenologous, enzymes) • Conservation of the reaction chemistry, but not the substrate specificity • Functional diversity in closely related proteins • Abundance of new structures

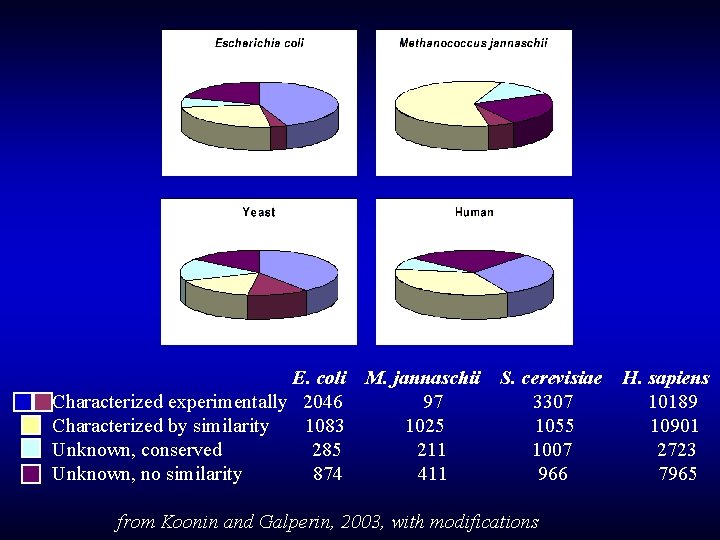
E. coli Characterized experimentally 2046 Characterized by similarity 1083 Unknown, conserved 285 Unknown, no similarity 874 M. jannaschii 97 1025 211 411 S. cerevisiae 3307 1055 1007 966 from Koonin and Galperin, 2003, with modifications H. sapiens 10189 10901 2723 7965

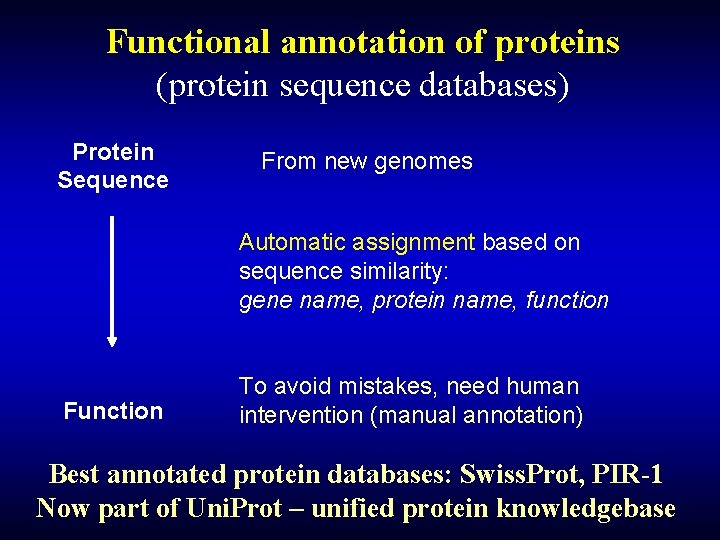
Functional annotation of proteins (protein sequence databases) Protein Sequence From new genomes Automatic assignment based on sequence similarity: gene name, protein name, function Function To avoid mistakes, need human intervention (manual annotation) Best annotated protein databases: Swiss. Prot, PIR-1 Now part of Uni. Prot – unified protein knowledgebase

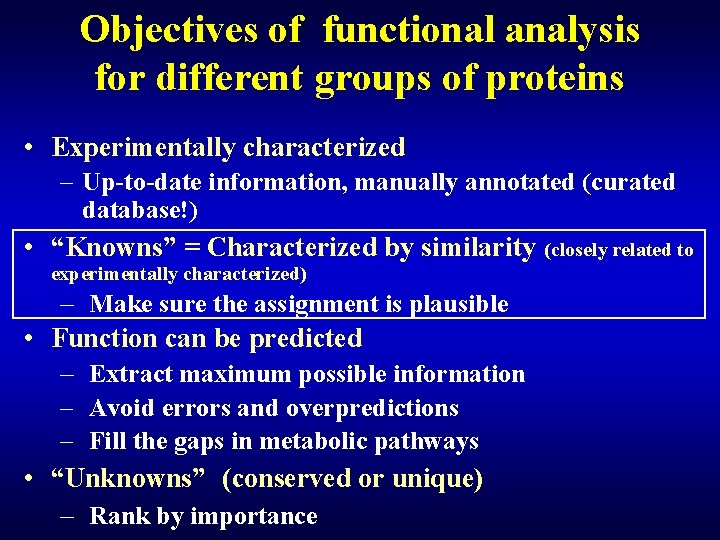
Objectives of functional analysis for different groups of proteins • Experimentally characterized – Up-to-date information, manually annotated (curated database!) • “Knowns” = Characterized by similarity (closely related to experimentally characterized) – Make sure the assignment is plausible • Function can be predicted – Extract maximum possible information – Avoid errors and overpredictions – Fill the gaps in metabolic pathways • “Unknowns” (conserved or unique) – Rank by importance

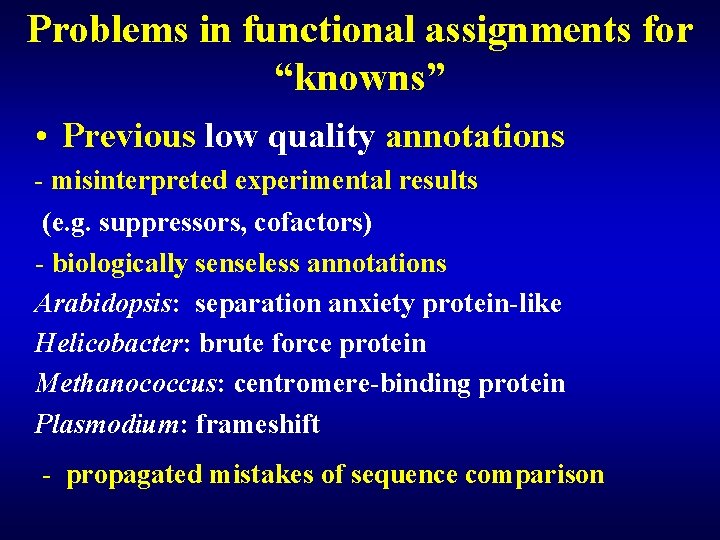
Problems in functional assignments for “knowns” • Previous low quality annotations - misinterpreted experimental results (e. g. suppressors, cofactors) - biologically senseless annotations Arabidopsis: separation anxiety protein-like Helicobacter: brute force protein Methanococcus: centromere-binding protein Plasmodium: frameshift - propagated mistakes of sequence comparison

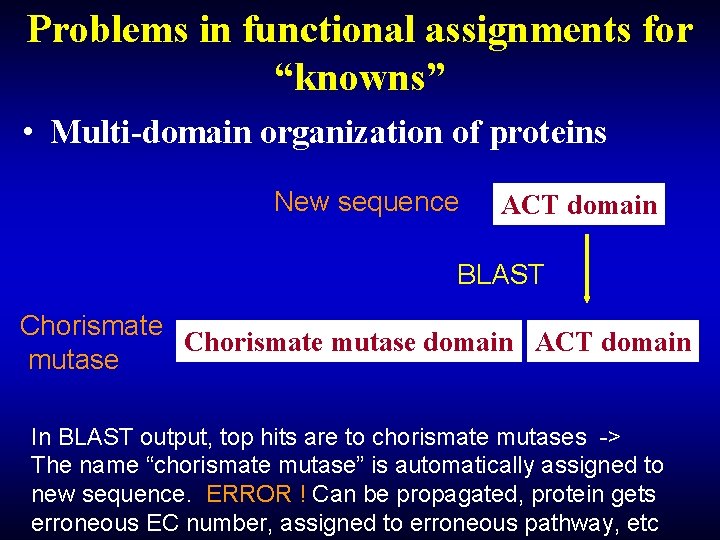
Problems in functional assignments for “knowns” • Multi-domain organization of proteins New sequence ACT domain BLAST Chorismate mutase domain ACT domain mutase In BLAST output, top hits are to chorismate mutases -> The name “chorismate mutase” is automatically assigned to new sequence. ERROR ! Can be propagated, protein gets erroneous EC number, assigned to erroneous pathway, etc

Problems in functional assignments for “knowns” • Low sequence complexity (coiled-coil, non-globular regions) • Non-orthologous gene displacement • Enzyme evolution (divergence in sequence and function)

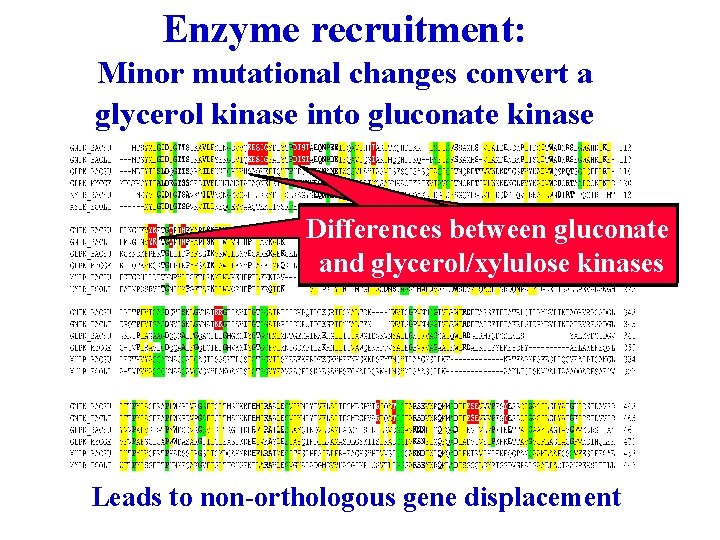
Enzyme recruitment: Minor mutational changes convert a glycerol kinase into gluconate kinase Differences between gluconate and glycerol/xylulose kinases Leads to non-orthologous gene displacement

Objectives of functional analysis for different groups of proteins • Experimentally characterized • “Knowns” = Characterized by similarity (closely related to experimentally characterized) – Make sure the assignment is plausible • Function can be predicted – Extract maximum possible information – Avoid errors and overpredictions – Fill the gaps in metabolic pathways • “Unknowns” (conserved or unique) – Rank by importance

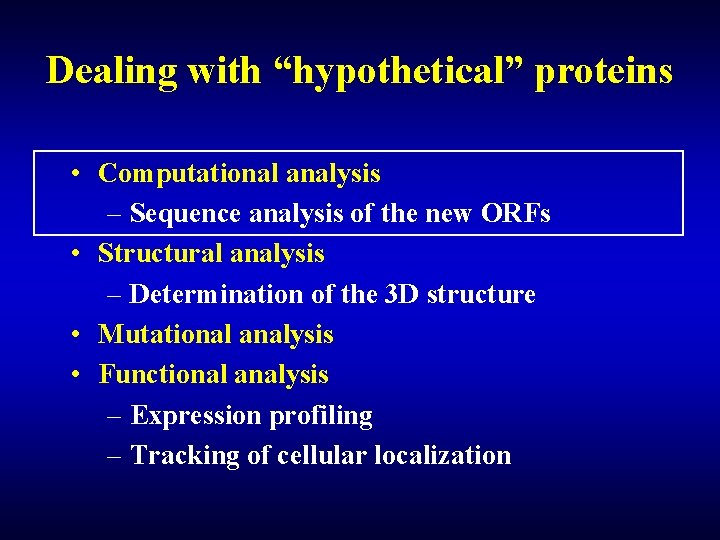
Dealing with “hypothetical” proteins • Computational analysis – Sequence analysis of the new ORFs • Structural analysis – Determination of the 3 D structure • Mutational analysis • Functional analysis – Expression profiling – Tracking of cellular localization

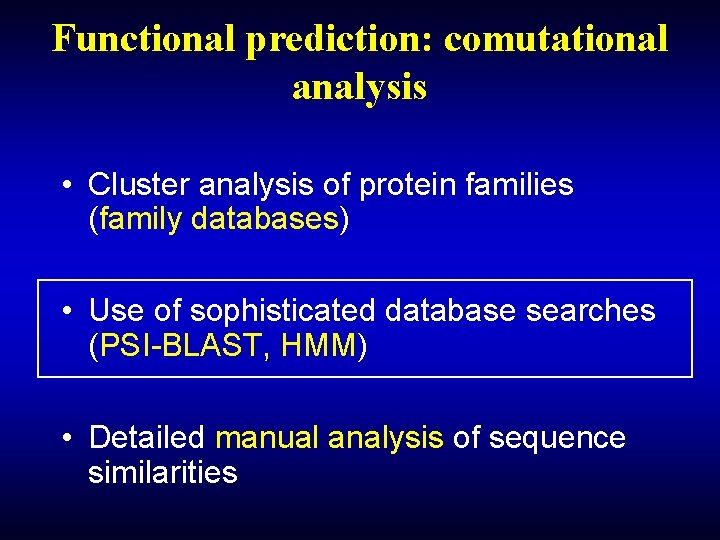
Functional prediction: comutational analysis • Cluster analysis of protein families (family databases) • Use of sophisticated database searches (PSI-BLAST, HMM) • Detailed manual analysis of sequence similarities

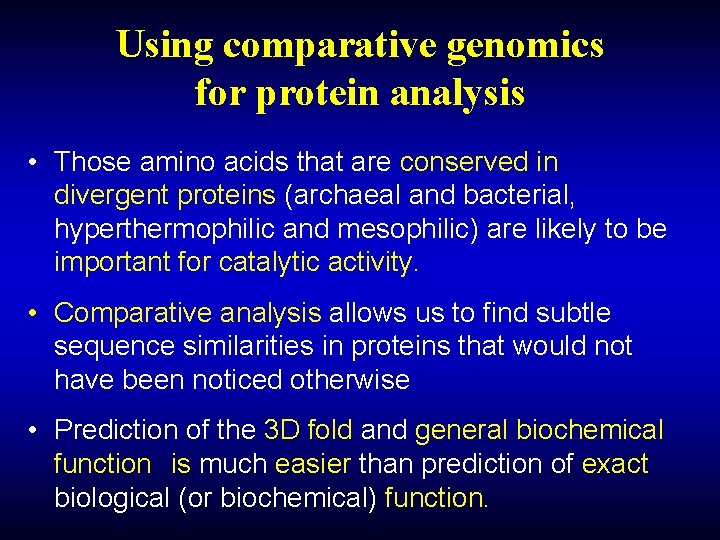
Using comparative genomics for protein analysis • Those amino acids that are conserved in divergent proteins (archaeal and bacterial, hyperthermophilic and mesophilic) are likely to be important for catalytic activity. • Comparative analysis allows us to find subtle sequence similarities in proteins that would not have been noticed otherwise • Prediction of the 3 D fold and general biochemical function is much easier than prediction of exact biological (or biochemical) function.

Using comparative genomics for protein analysis • Reaction chemistry often remains conserved even when sequence diverges almost beyond recognition • Sequence database searches that use exotic or highly divergent query sequences often reveal more subtle relationships than those using queries from humans or standard model organisms (E. coli, yeast, worm, fly). • Sequence analysis complements structural comparisons and can greatly benefit from them

Poorly characterized protein families • Enzyme activity can be predicted, the substrate remains unknown (ATPases, GTPases, oxidoreductases, methyltransferases, acetyltransferases) • Helix-turn-helix motif proteins (predicted transcriptional regulators) • Membrane transporters

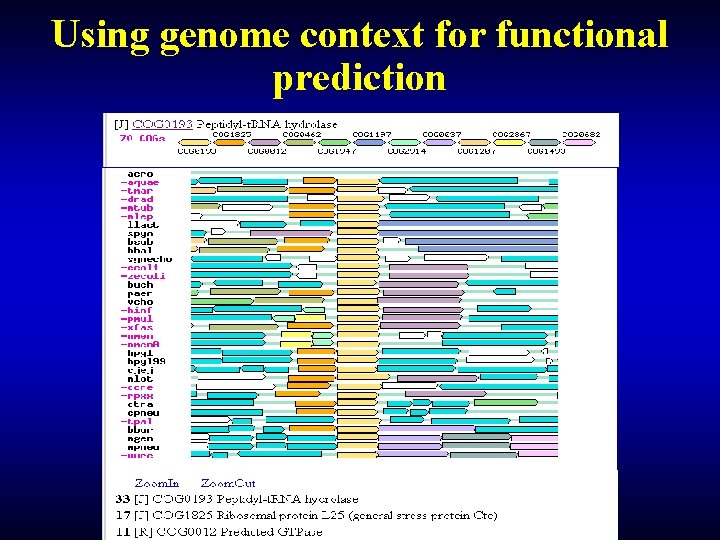
Functional prediction: computational analysis • Phylogenetic distribution – Wide - most likely essential – Narrow - probably clade-specific – Patchy - most intriguing, niche-specific • Domain association – Rosetta Stone (for multidomain proteins) • Gene neighborhood (operon organization)

Using genome context for functional prediction

Functional Prediction: Role of Structural Genomics Protein Structure Initiative: Determine 3 D Structures of All Proteins – Family Classification: Organize Protein Sequences into Families, collect families without known structures – Target Selection: Select Family Representatives as Targets – Structure Determination: X-Ray Crystallography or NMR Spectroscopy – Homology Modeling: Build Models for Other Proteins by Homology – Functional prediction based on structure

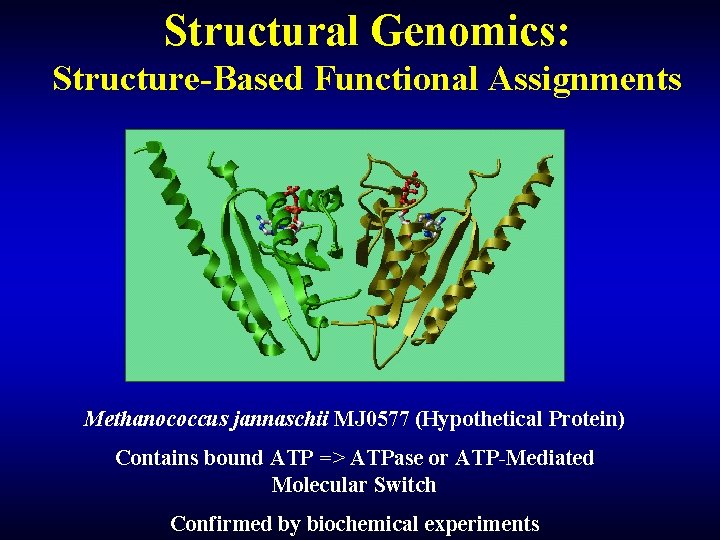
Structural Genomics: Structure-Based Functional Assignments Methanococcus jannaschii MJ 0577 (Hypothetical Protein) Contains bound ATP => ATPase or ATP-Mediated Molecular Switch Confirmed by biochemical experiments

Crystal structure is not a function!

Functional prediction: problem areas • Identification of protein-coding regions • Delineation of potential function(s) for distant paralogs • Identification of domains in the absense of close homologs • Analysis of proteins with low sequence complexity

Objectives of functional analysis for different groups of proteins • Experimentally characterized – Up-to-date information, manually annotated • “Knowns” = Characterized by similarity (closely related to experimentally characterized) – Make sure the assignment is plausible • Function can be predicted – Extract maximum possible information – Avoid errors and overpredictions – Fill the gaps in metabolic pathways • “Unknowns” (conserved or unique) – Rank by importance

“Unknown unknowns” • Phylogenetic distribution – Wide - most likely essential – Narrow - probably clade-specific – Patchy - most intriguing, nichespecific

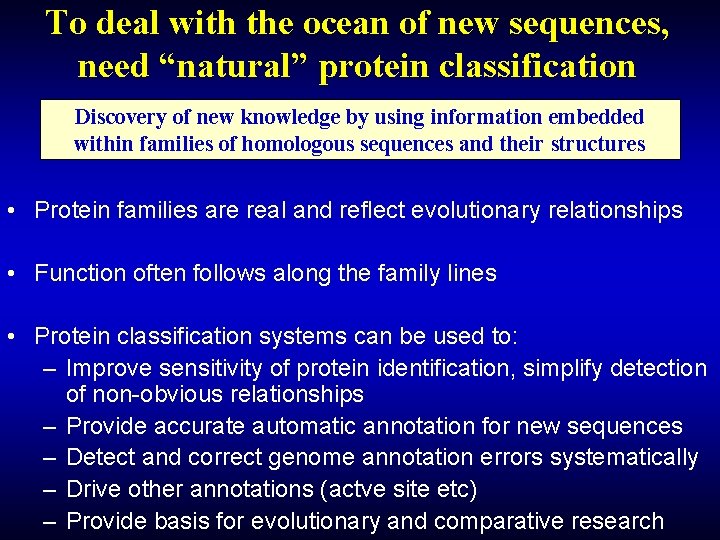
To deal with the ocean of new sequences, need “natural” protein classification Discovery of new knowledge by using information embedded within families of homologous sequences and their structures • Protein families are real and reflect evolutionary relationships • Function often follows along the family lines • Protein classification systems can be used to: – Improve sensitivity of protein identification, simplify detection of non-obvious relationships – Provide accurate automatic annotation for new sequences – Detect and correct genome annotation errors systematically – Drive other annotations (actve site etc) – Provide basis for evolutionary and comparative research

The ideal system would be: • Comprehensive, with each sequence classified either as a member of a family or as an “orphan” sequence • Hierarchical and based on evolution, with families united into superfamilies on the basis of distant homology • Allow for simultaneous use of the whole protein and domain information (domains mapped onto proteins) • Allow for automatic classification/annotation of new sequences • Expertly curated (family name, function, evidence attribution (experimental vs predicted), background etc). This is the only way to avoid annotation errors and prevent error propagation

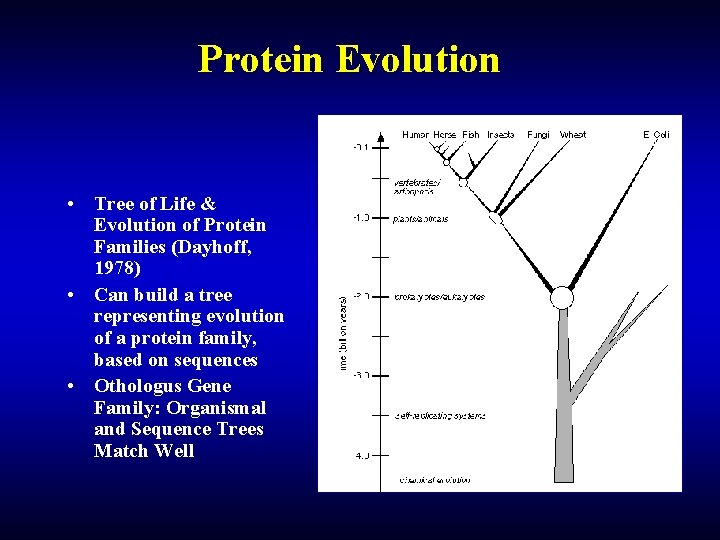
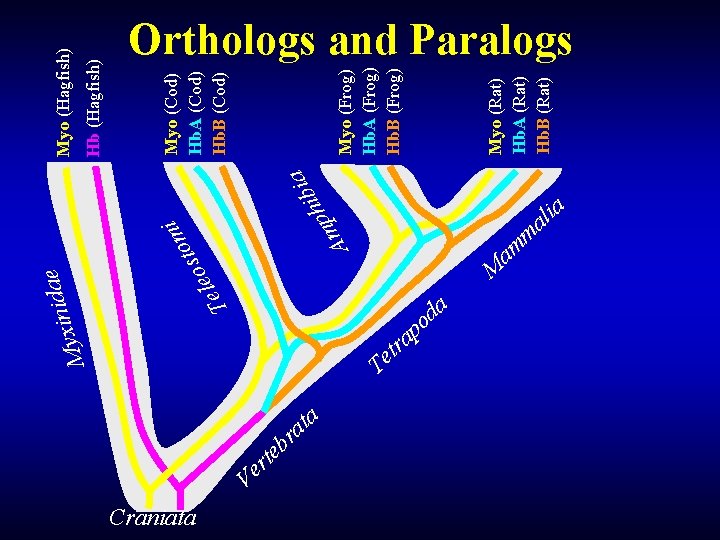
Protein Evolution • Tree of Life & Evolution of Protein Families (Dayhoff, 1978) • Can build a tree representing evolution of a protein family, based on sequences • Othologus Gene Family: Organismal and Sequence Trees Match Well

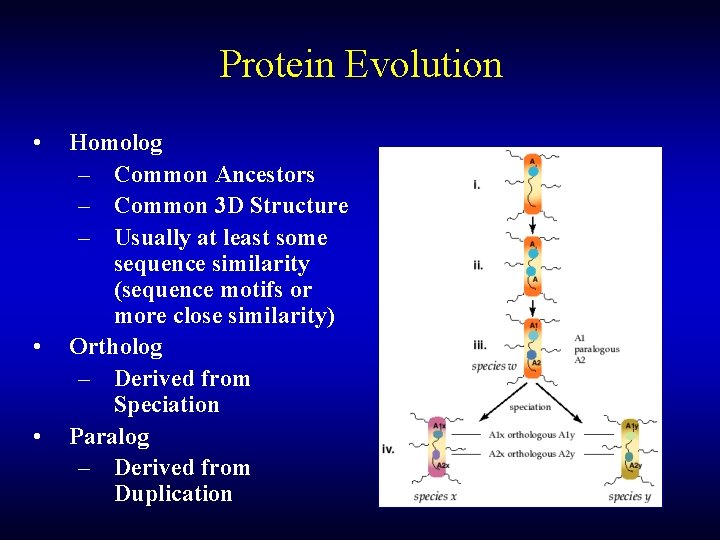
Protein Evolution • • • Homolog – Common Ancestors – Common 3 D Structure – Usually at least some sequence similarity (sequence motifs or more close similarity) Ortholog – Derived from Speciation Paralog – Derived from Duplication

M T Craniata a i b i h p m A i m sto o e l e e a d i n yxi t e T r t a r b tr e Ve a Myo (Rat) Hb. A (Rat) Hb. B (Rat) Myo (Frog) Hb. A (Frog) Hb. B (Frog) Myo (Cod) Hb. A (Cod) Hb. B (Cod) Hb (Hagfish) Myo (Hagfish) Orthologs and Paralogs M am m a d o p a ia l a

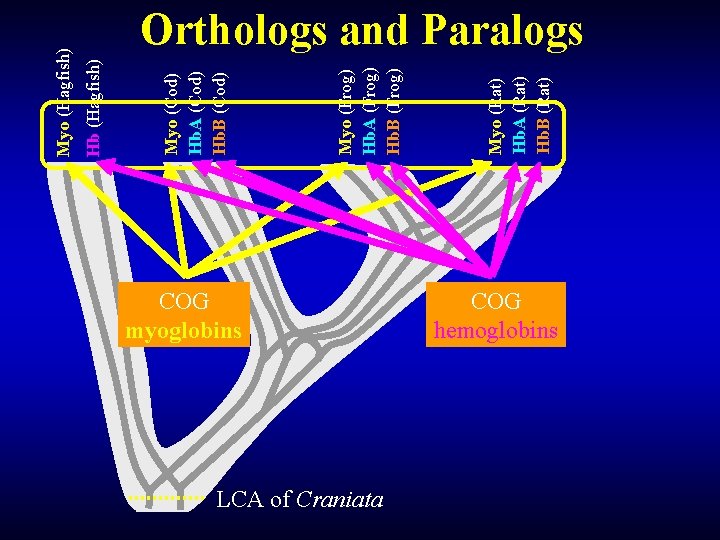
COG myoglobins LCA of Craniata Myo (Rat) Hb. A (Rat) Hb. B (Rat) Myo (Frog) Hb. A (Frog) Hb. B (Frog) Myo (Cod) Hb. A (Cod) Hb. B (Cod) Hb (Hagfish) Myo (Hagfish) Orthologs and Paralogs COG hemoglobins

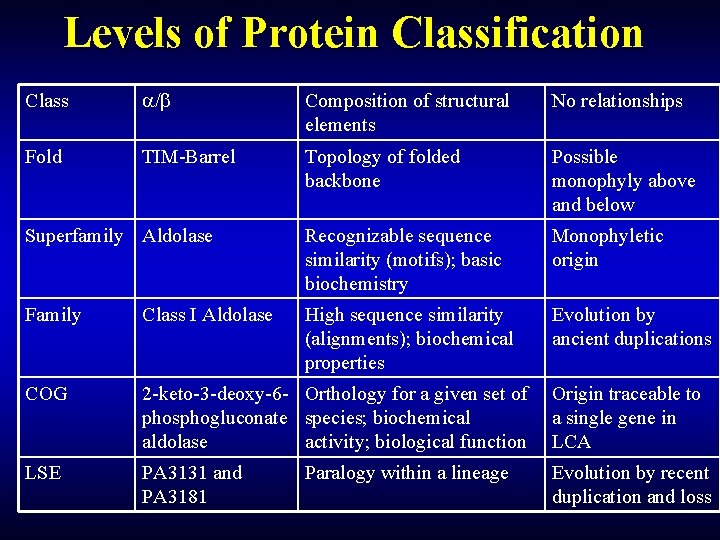
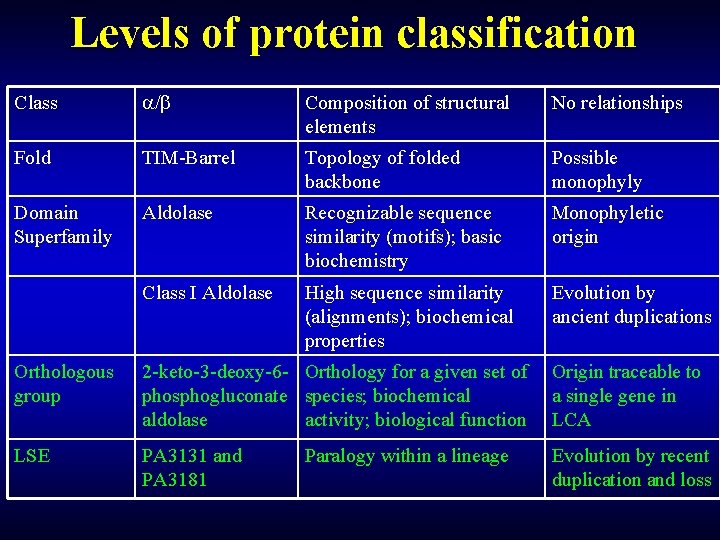
Levels of Protein Classification Class / Composition of structural elements No relationships Fold TIM-Barrel Topology of folded backbone Possible monophyly above and below Superfamily Aldolase Recognizable sequence similarity (motifs); basic biochemistry Monophyletic origin Family Class I Aldolase High sequence similarity (alignments); biochemical properties Evolution by ancient duplications COG 2 -keto-3 -deoxy-6 - Orthology for a given set of phosphogluconate species; biochemical aldolase activity; biological function Origin traceable to a single gene in LCA LSE PA 3131 and PA 3181 Evolution by recent duplication and loss Paralogy within a lineage

Protein Family-Domain-Motif • Domain: Evolutionary/Functional/Structural Unit Domain = structurally compact, independently folding unit that forms a stable three-dimentional structure and shows a certain level of evolutionary conservation. Usually, corresponds to an evolutionary unit. A protein can consist of a single domain or multiple domains. Proteins have modular structure. • Motif: Conserved Functional/Structural Site

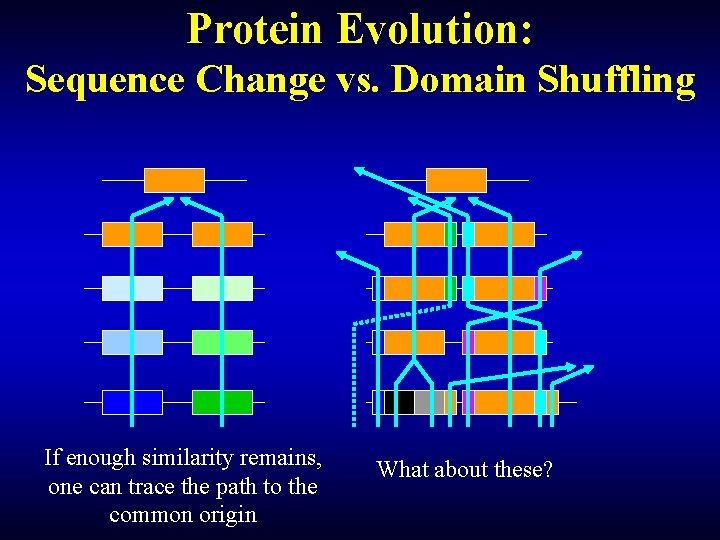
Protein Evolution: Sequence Change vs. Domain Shuffling If enough similarity remains, one can trace the path to the common origin What about these?

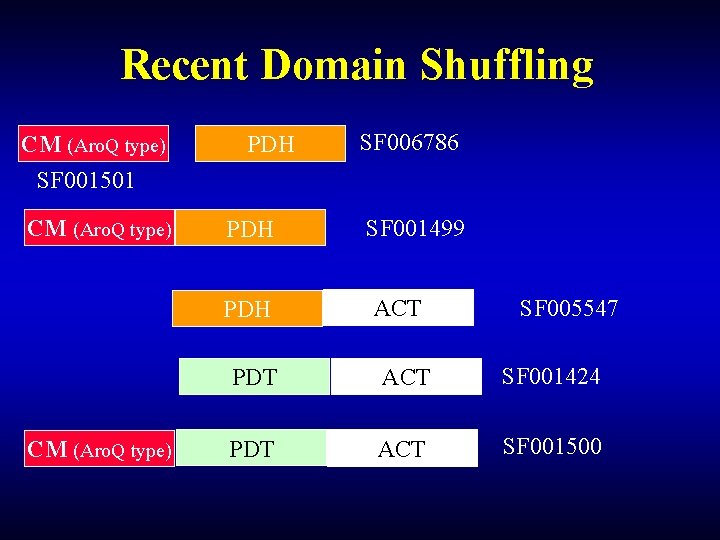
Recent Domain Shuffling CM (Aro. Q type) SF 001501 CM (Aro. Q type) PDH PDH CM (Aro. Q type) SF 006786 SF 001499 ACT SF 005547 PDT ACT SF 001424 PDT ACT SF 001500

Practical classification of proteins: setting realistic goals We strive to reconstruct the natural classification of proteins to the fullest possible extent BUT Domain shuffling rapidly degrades the continuity in the protein structure (faster than sequence divergence degrades similarity) THUS The further we extend the classification, the finer is the domain structure we need to consider SO We need to compromise between the depth of analysis and protein integrity OR … Credit: Dr. Y. Wolf, NCBI

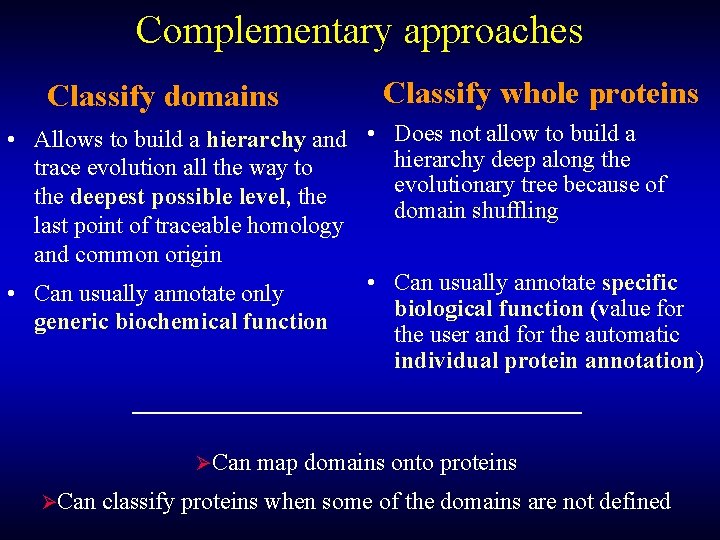
Complementary approaches Classify domains Classify whole proteins • Allows to build a hierarchy and • Does not allow to build a hierarchy deep along the trace evolution all the way to evolutionary tree because of the deepest possible level, the domain shuffling last point of traceable homology and common origin • Can usually annotate specific • Can usually annotate only biological function (value for generic biochemical function the user and for the automatic individual protein annotation) ØCan map domains onto proteins ØCan classify proteins when some of the domains are not defined

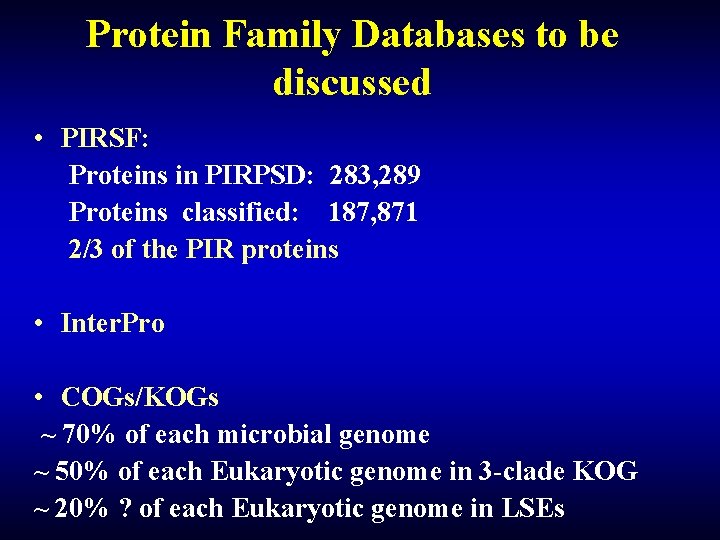
Protein Family Databases to be discussed • PIRSF: Proteins in PIRPSD: 283, 289 Proteins classified: 187, 871 2/3 of the PIR proteins • Inter. Pro • COGs/KOGs ~ 70% of each microbial genome ~ 50% of each Eukaryotic genome in 3 -clade KOG ~ 20% ? of each Eukaryotic genome in LSEs

PIR Web Site (http: //pir. georgetown. edu)

PIRSF protein classification system ØBasic concept: l. A network classification system based on evolutionary relationship of whole proteins ØBasic unit = PIRSF Family l. Homeomorphic (end-to-end similarity with common domain architecture) l. Monophyletic (common ancestry) ØDomains and motifs are mapped onto PIRSF
![Whole protein functional annotation Ni Fehydrogenase maturation factor carbamoyl phosphateconverting enzyme Acylphosphatase Znf Whole protein functional annotation [Ni. Fe]-hydrogenase maturation factor, carbamoyl phosphate-converting enzyme Acylphosphatase – Znf](https://slidetodoc.com/presentation_image_h2/19561593889fc34fbf9f79bcece9e9a2/image-47.jpg)
Whole protein functional annotation [Ni. Fe]-hydrogenase maturation factor, carbamoyl phosphate-converting enzyme Acylphosphatase – Znf x 2 – Yrd. C - SF 006256 related to Peptidase M 22 On the basis of domain composition alone: - can not tell that this is a hydrogenase maturation factor - various speculations for exact role: RNA-binding translation factor, maturation protease …. Recent data: carbamoyl phosphate-converting enzyme

PIRSF classification is based on evolution • Sequence similarity, domain architecture and phyletic pattern guide PIRSF creation/curation • PIRSF Families range from ancient conserved proteins to unique lineage specific expansions • PIRSF Families are monophyletic (with some exceptions) • Members are homologs (may be orthologs or paralogs)

Levels of protein classification Class / Composition of structural elements No relationships Fold TIM-Barrel Topology of folded backbone Possible monophyly Domain Superfamily Aldolase Recognizable sequence similarity (motifs); basic biochemistry Monophyletic origin Class I Aldolase High sequence similarity (alignments); biochemical properties Evolution by ancient duplications Orthologous group 2 -keto-3 -deoxy-6 - Orthology for a given set of phosphogluconate species; biochemical aldolase activity; biological function Origin traceable to a single gene in LCA LSE PA 3131 and PA 3181 Evolution by recent duplication and loss Paralogy within a lineage

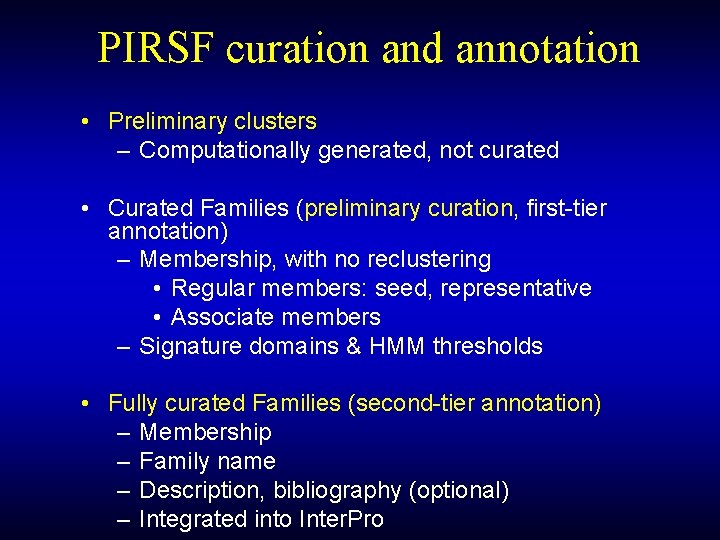
PIRSF curation and annotation • Preliminary clusters – Computationally generated, not curated • Curated Families (preliminary curation, first-tier annotation) – Membership, with no reclustering • Regular members: seed, representative • Associate members – Signature domains & HMM thresholds • Fully curated Families (second-tier annotation) – Membership – Family name – Description, bibliography (optional) – Integrated into Inter. Pro

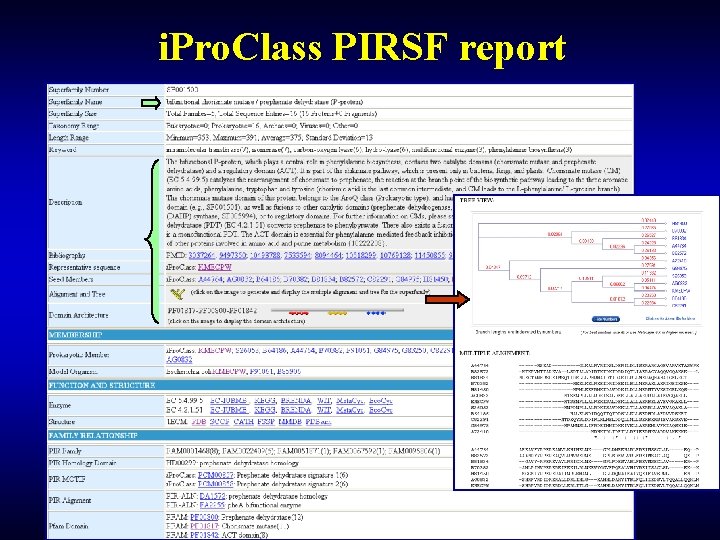
i. Pro. Class PIRSF report

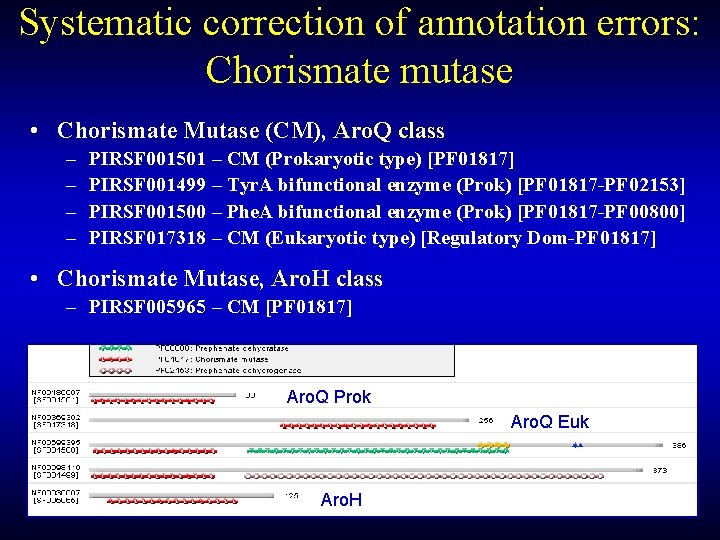
Systematic correction of annotation errors: Chorismate mutase • Chorismate Mutase (CM), Aro. Q class – – PIRSF 001501 – CM (Prokaryotic type) [PF 01817] PIRSF 001499 – Tyr. A bifunctional enzyme (Prok) [PF 01817 -PF 02153] PIRSF 001500 – Phe. A bifunctional enzyme (Prok) [PF 01817 -PF 00800] PIRSF 017318 – CM (Eukaryotic type) [Regulatory Dom-PF 01817] • Chorismate Mutase, Aro. H class – PIRSF 005965 – CM [PF 01817] Aro. Q Prok Aro. Q Euk Aro. H

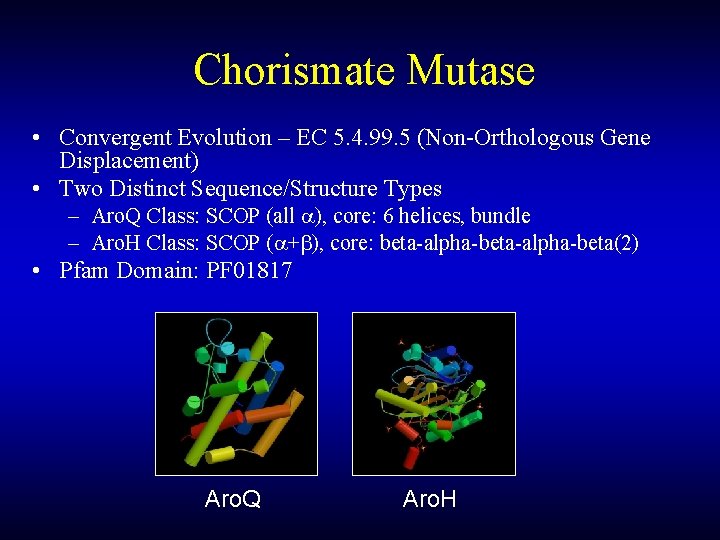
Chorismate Mutase • Convergent Evolution – EC 5. 4. 99. 5 (Non-Orthologous Gene Displacement) • Two Distinct Sequence/Structure Types – Aro. Q Class: SCOP (all ), core: 6 helices, bundle – Aro. H Class: SCOP ( + ), core: beta-alpha-beta(2) • Pfam Domain: PF 01817 Aro. Q Aro. H

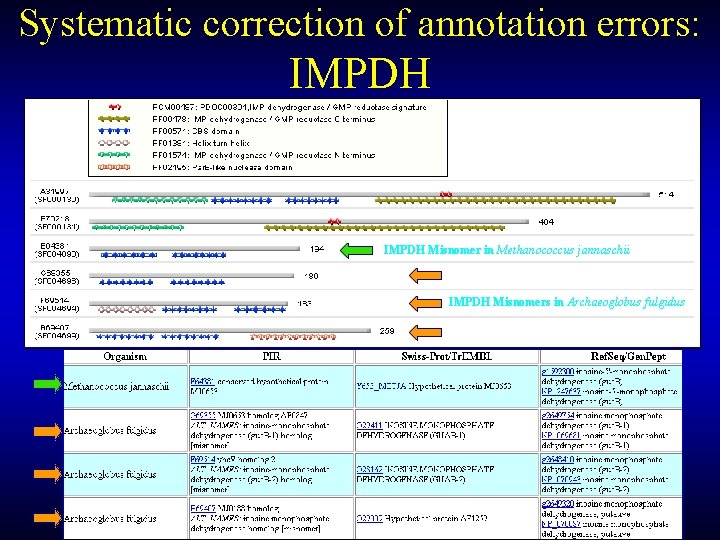
Systematic correction of annotation errors: IMPDH Misnomer in Methanococcus jannaschii IMPDH Misnomers in Archaeoglobus fulgidus

Propagation of protein annotation within Uni. Prot (under development) • Homeomorphic family level is the primary PIRSF curation and annotation level, most invested with biological meaning • Reliable automatic assignment of new family members • Systematic detection of annotation errors (curator looks at every protein annotation in the family) • PIRSF Family/Subfamily name - can be applied to every member - make possible the automatic transfer of name from PIRSF to every unnamed/unannotated member in Uni. Prot • Preventing error propagation: evidence attribution experimental (validated and tentative) and predicted

Inter. Pro (at EBI) -Inter. Pro is an integrated resource for protein families, domains and sites. - Inter. Pro combines a number of databases that use different methodologies. By uniting the member databases, Inter. Pro capitalizes on their individual strengths, producing a powerful integrated diagnostic tool. Member databases: PROSITE, PRINTS, Pfam, SMART, Pro. Dom, and TIGRFAMs PIR to be added soon SWISSPROT and Tr. EMBL matches used as examples

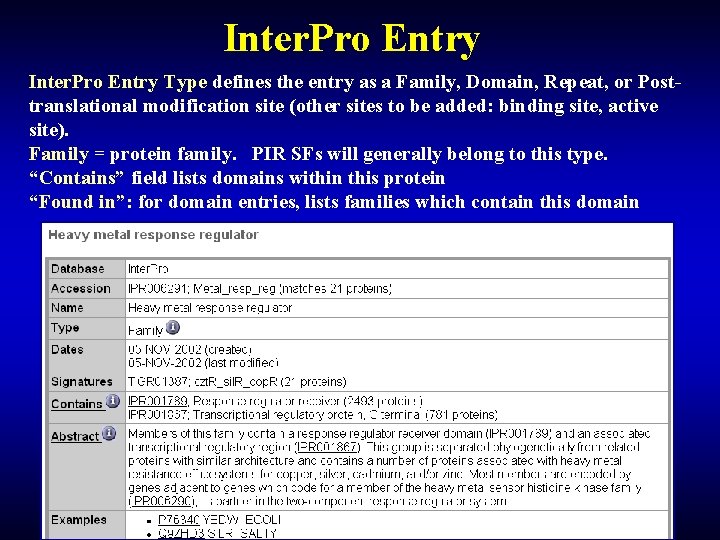
Inter. Pro Entry Type defines the entry as a Family, Domain, Repeat, or Posttranslational modification site (other sites to be added: binding site, active site). Family = protein family. PIR SFs will generally belong to this type. “Contains” field lists domains within this protein “Found in”: for domain entries, lists families which contain this domain

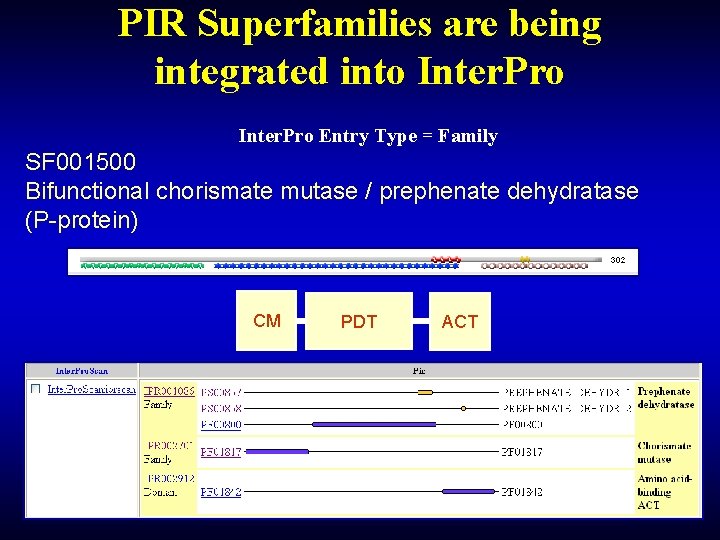
PIR Superfamilies are being integrated into Inter. Pro Entry Type = Family SF 001500 Bifunctional chorismate mutase / prephenate dehydratase (P-protein) CM PDT ACT

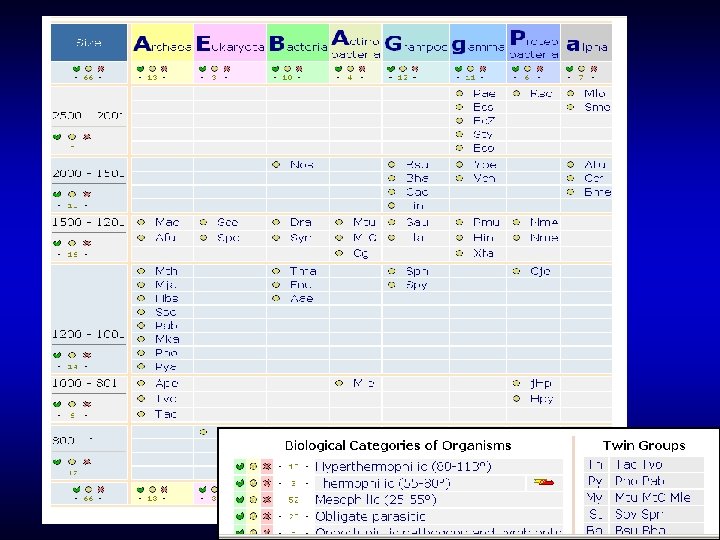
COGs (Clusters of Orthologous Groups) (at NCBI) • Complete genomes • Reciprocal best hits • No score cutoffs Comparative genomics – a branch of computational biology that uses complete genome sequences

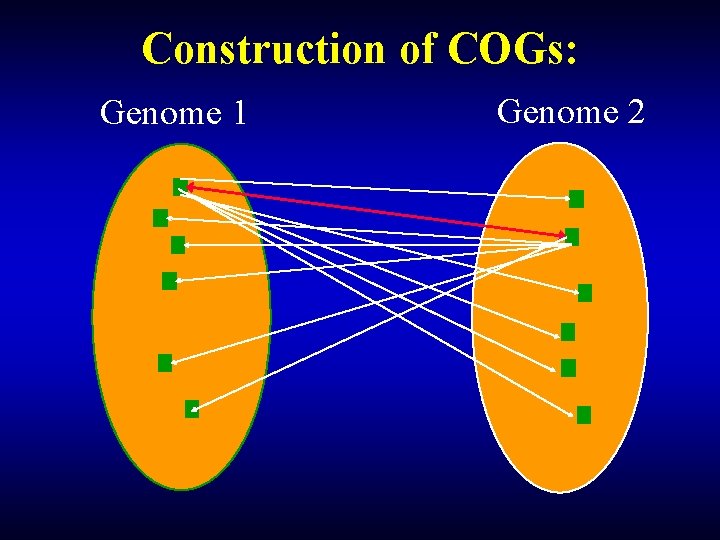
Construction of COGs: Genome 1 Genome 2

Construction of COGs: Bidirectional best hit E. coli fbp Yeast YLR 377 c Triangle Bidirectional the simplest best hit COG Bidirectional best hit Synechocystis slr 0952

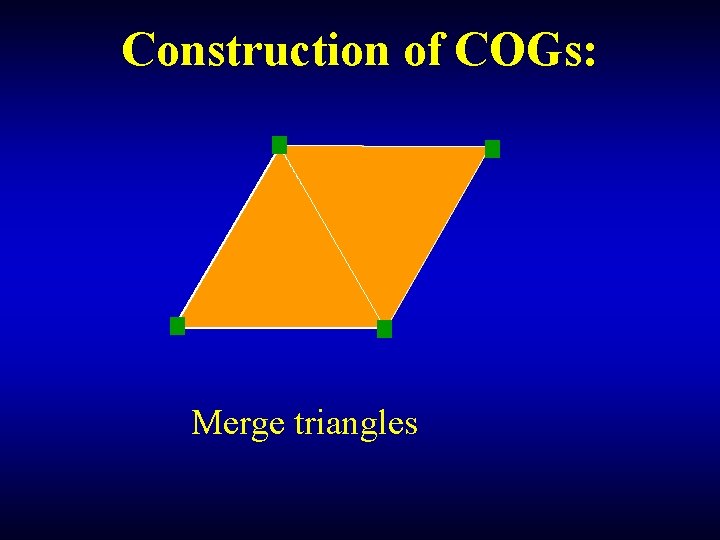
Construction of COGs: Merge triangles

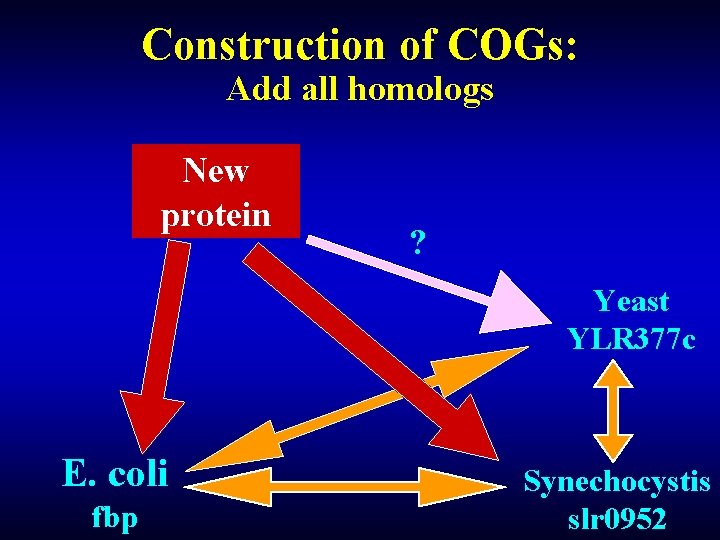
Construction of COGs: Add all homologs New protein ? Yeast YLR 377 c E. coli fbp Synechocystis slr 0952




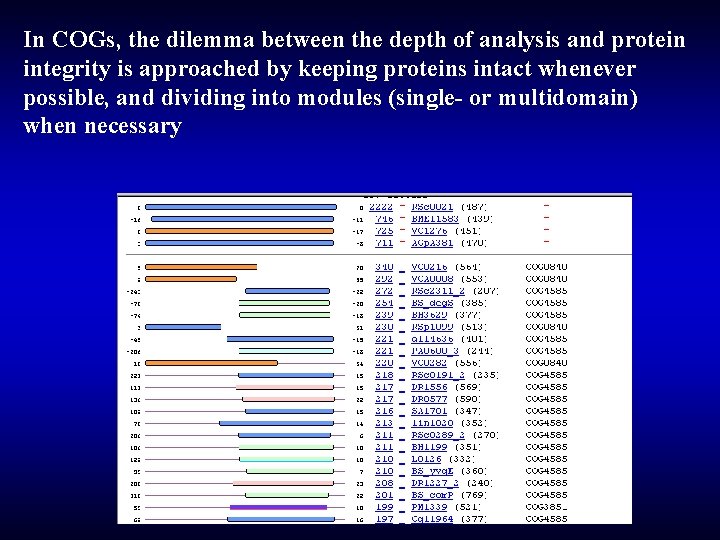
In COGs, the dilemma between the depth of analysis and protein integrity is approached by keeping proteins intact whenever possible, and dividing into modules (single- or multidomain) when necessary

Impact of genomics • Single protein level – Discovery of new enzymes and superfamilies – Prediction of active sites and 3 D structures • Pathway level – Identification of “missing” enzymes – Prediction of alternative enzyme forms – Identification of potential drug targets • Cellular metabolism level – Multisubunit protein systems – Membrane energy transducers – Cellular signaling systems

What to do with a new porotein sequence • I. Basic: - Domain analysis (SMART = recommended; PFAM, CDD is included in BLAST output) - BLAST - Curated protein family databases (PIRSF, Inter. Pro, COGs) • II. If not sufficient: - Literature (Pub. Med) from links from individual entries on the BLAST output (look for Swiss. Prot entries first) - Refined Pub. Med search using gene/protein names, synomims, function and other terms you found • III. Advanced: - Multiple sequence alignments - Phylogenetic tree reconstruction

Case Study: Prediction verified: GGDEF domain • Proteins containing this domain: Caulobacter crescentus Ple. D controls swarmer cell - stalk cell transition (Hecht and Newton, 1995). In Rhizobium leguminosarum, Acetobacter xylinum, required for cellulose biosynthesis (regulation) • Predicted to be involved in signal transduction because it is found in fusions with other signaling domains (receiver, etc) • In Acetobacter xylinum, cyclic di-GMP is a specific nucleotide regulator of cellulose synthase (signalling molecule). Multidomain protein with GGDEF domain was shown to have diguanylate cyclase activity (Tal et al. , 1998) • Detailed sequence analysis tentatively predicts GGDEF to be a diguanylate cyclase domain (Pei and Grishin, 2001) • Complementation experiments prove diguanylate cyclase activity of GGDEF (Ausmees et al. , 2001)

Examples for analysis: 1. Retrieve one of the following protein sequences: PIR: C 69086 D 64376 Gen. Bank GI: 15679635. Using analysis tools available on the web, check if the functional annotation is correct, and provide correct annotation without looking at internal PIR or COG annotations (Run BLAST with CDsearch and SMART to start with). When you are done, look at the PIR curated SF annotation, and at COG annotations. What caused the wrong annotations? In BLAST outputs for these sequences, do you see other wrongly annotated proteins (same and different type errors)? • Next, analyze the C-terminal domain of these proteins by PSI-BLAST (and alignment analysis) and suggest any speculations as to its function (homework).

Examples for analysis: 2. • Retrieve the following sequence: GI: 7019521 • Take a look at the associated publication (reference). • Analyze the sequence to see if any additional information can be obtained (run PSI-BLAST, and (as a homework) construct multiple alignment). • Take a look at taxonomy report: what does it tell you? • Find experimental paper associated with one of the sequences found by PSI-BLAST. What annotation is appropriate for this sequence and for the entire family?

Examples for analysis: 3. Predict the function of the following proteins: • Gen. Bank: GI: 27716853 • E. coli Yje. E protein Verify and/or correct the following functional annotations. Can you explain why the erroneous annotations were made? • PIR: H 87387 • Gen. Bank: GI: 15606003 GI: 15807219 • PIR: F 70338

Examples for analysis: 4. Homework: an exercise in transitive relationships: Start with >gi|20093648|ref|NP_613495. 1| Uncharacterized membrane protein, conserved in Archaea [Methanopyrus kandleri AV 19] (this is a short membrane protein); run PSI-BLAST, make sure you have filtering, complexity and CD-search off. There are no good hits but a bunch of sub-threshold ones. Collect "suspect" relations, use them as queries and expand the net. You will be able to come up with two proteins: >gi|21227474|ref|NP_633396. 1| hypothetical protein [Methanosarcina mazei Goe 1] and >gi|14324537|dbj|BAB 59464. 1| hypothetical protein [Thermoplasma volcanium] When used as a PSI-BLAST query, the first will tie the Methanopyrus protein into a group, while the second will tie this group to the Sec 61 subunit of preprotein translocase. Then, of course, you can obtain the same result with CD-search in a single step .