From gene to phenotype genotype transcription and translation



- Slides: 69

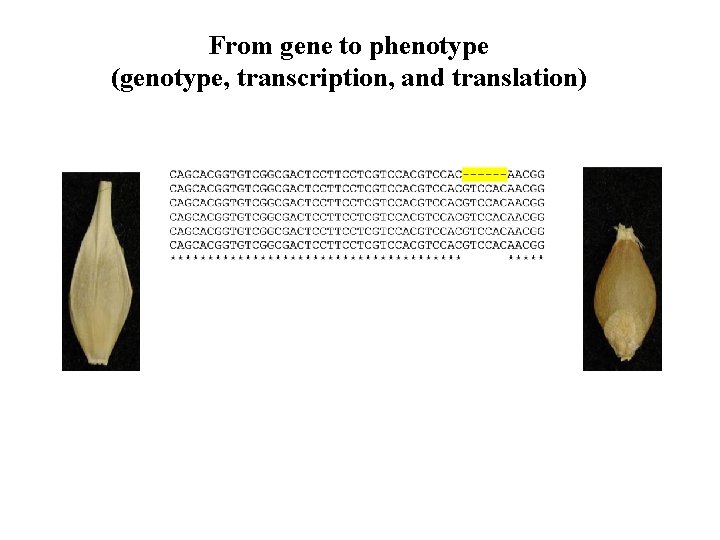
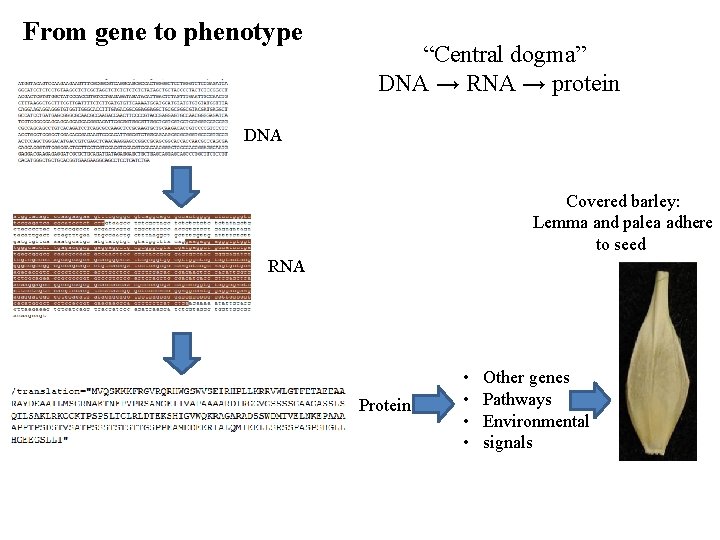
From gene to phenotype (genotype, transcription, and translation)

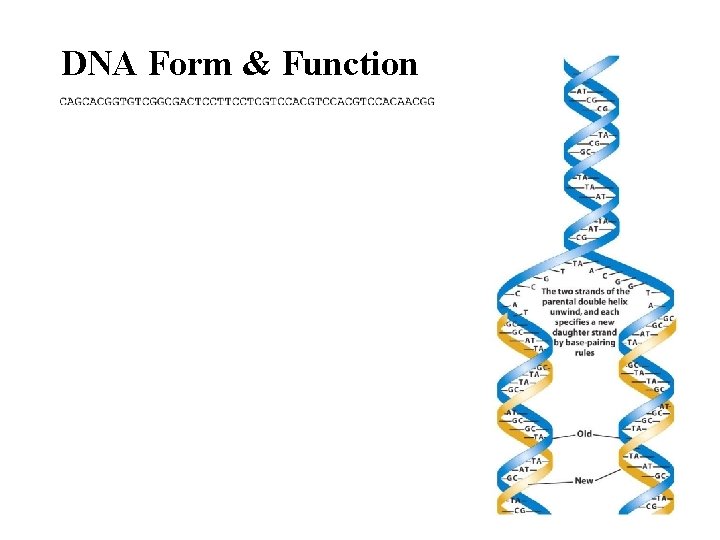
DNA Form & Function

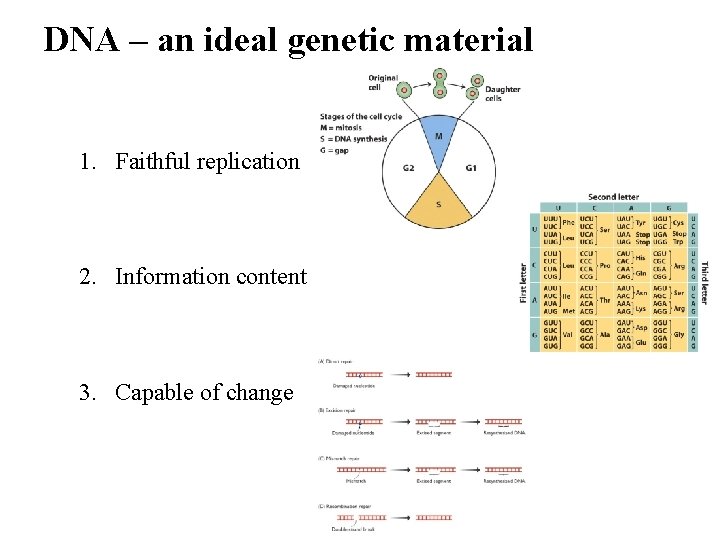
DNA – an ideal genetic material 1. Faithful replication 2. Information content 3. Capable of change

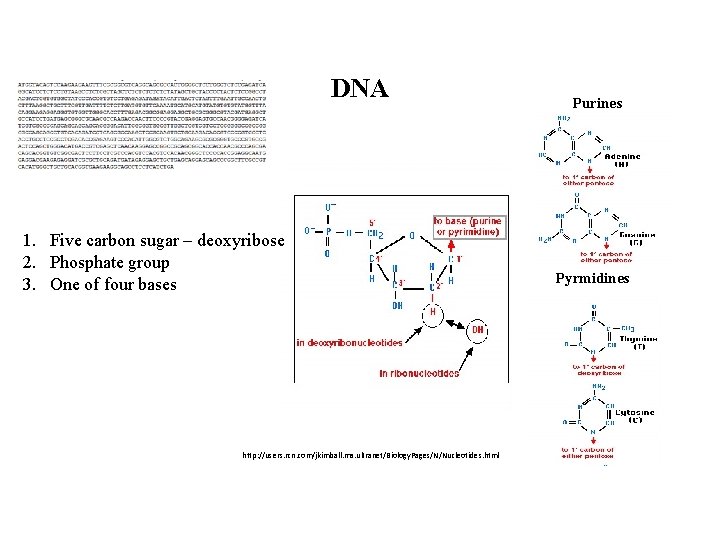
DNA 1. Five carbon sugar – deoxyribose 2. Phosphate group 3. One of four bases http: //users. rcn. com/jkimball. ma. ultranet/Biology. Pages/N/Nucleotides. html Purines Pyrmidines

DNA: Polymerization Nucleotide + …… http: //users. rcn. com/jkimball. ma. ultranet/Biology. Pages/N/Nucleotides. ht ml

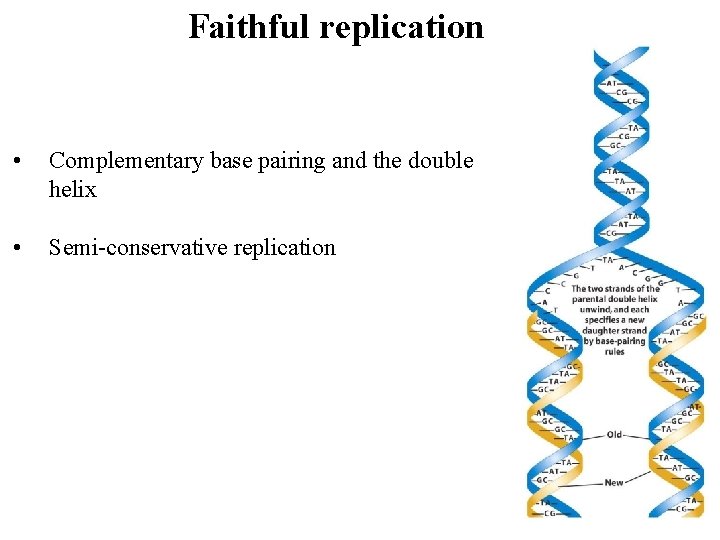
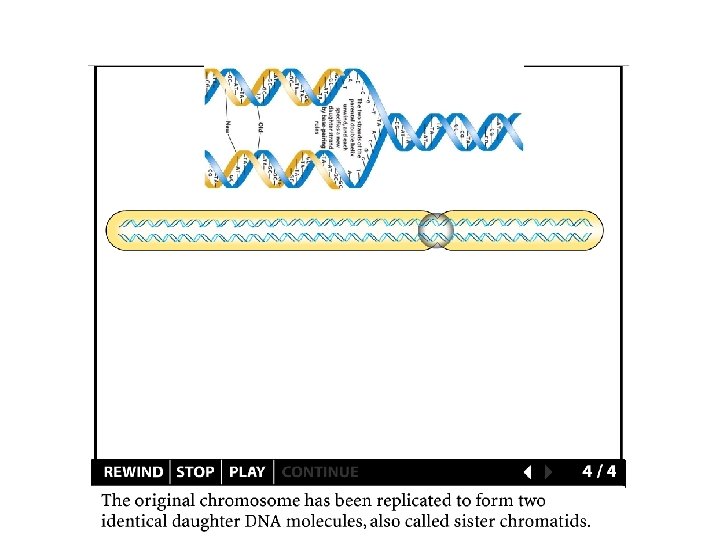
Faithful replication • Complementary base pairing and the double helix • Semi-conservative replication

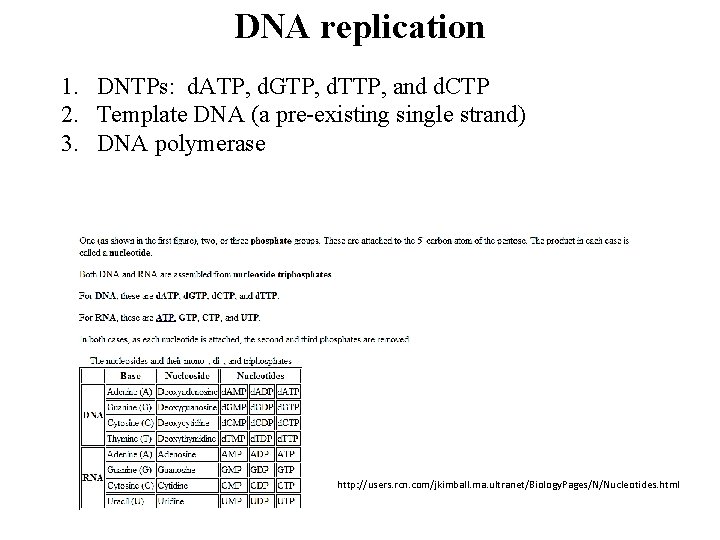
DNA replication 1. DNTPs: d. ATP, d. GTP, d. TTP, and d. CTP 2. Template DNA (a pre-existing single strand) 3. DNA polymerase http: //users. rcn. com/jkimball. ma. ultranet/Biology. Pages/N/Nucleotides. html

DNA replication Template DNA (a pre-existing single strand)

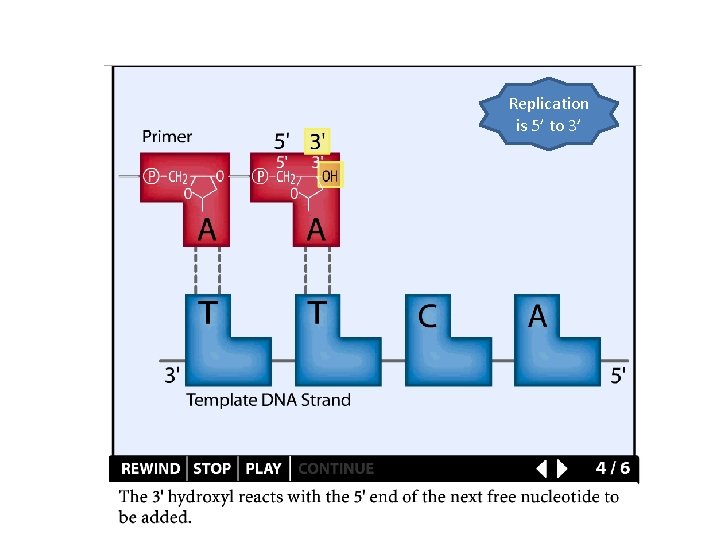
DNA replication DNA Polymerases catalyze the formation of a phosphodiester bond between the 3'-OH of the deoxyribose on the last nucleotide and the 5' phosphate of the d. NTP precursor

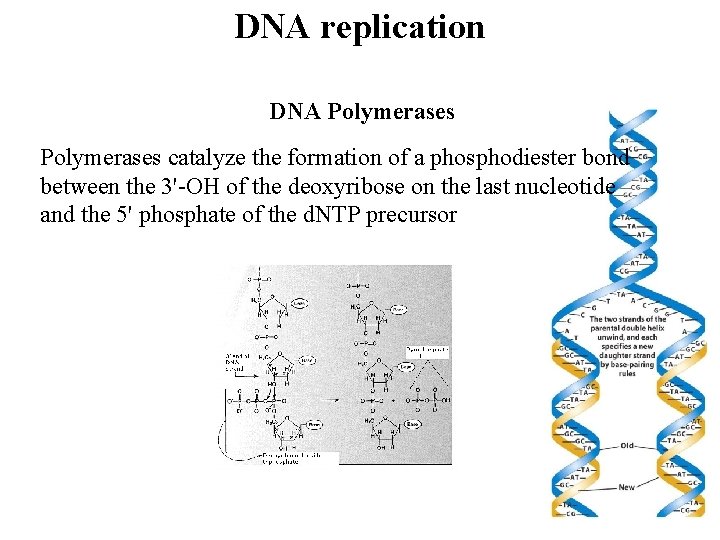
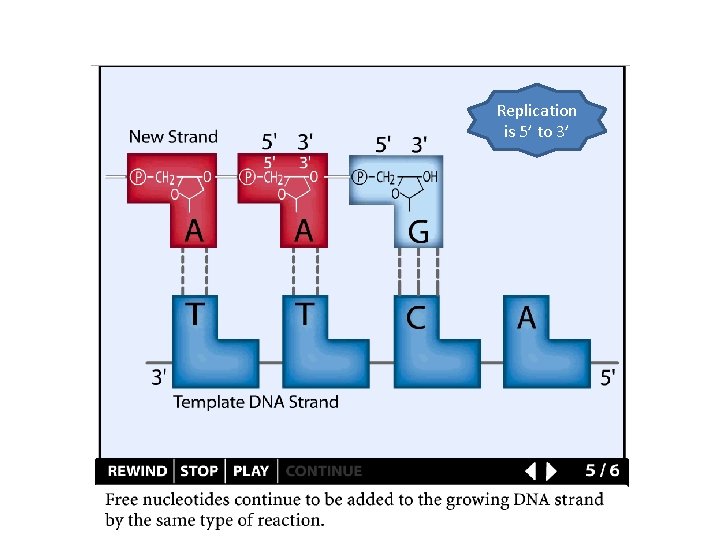
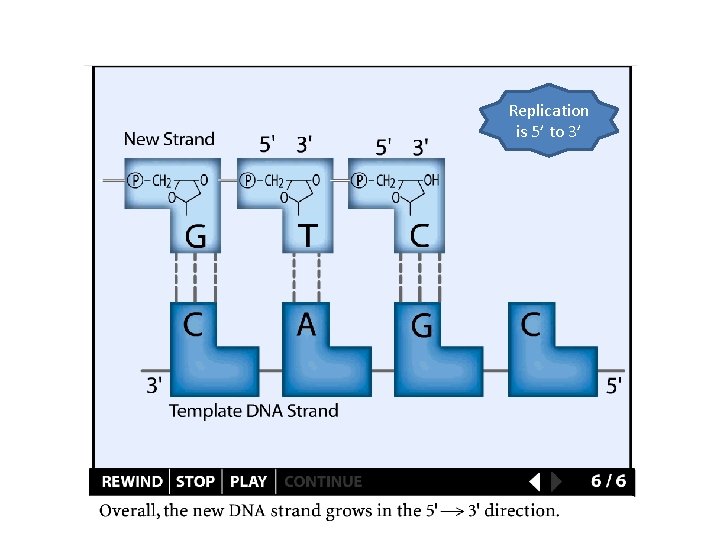
Replication is 5’ to 3’

Replication is 5’ to 3’

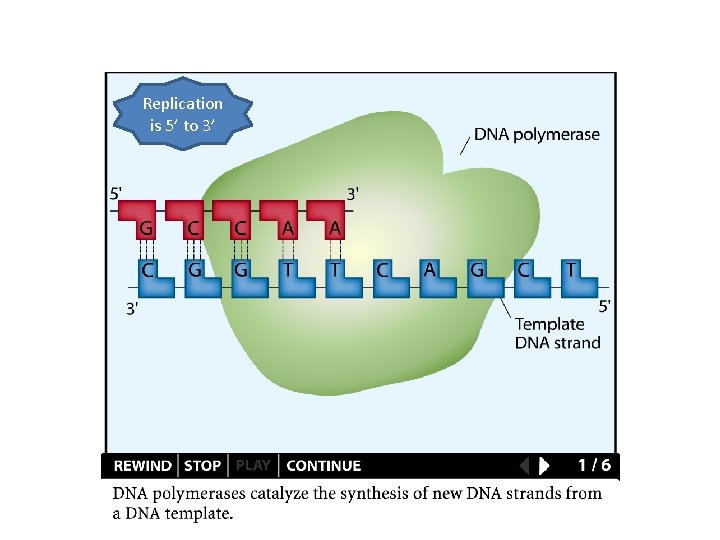
Replication is 5’ to 3’

Replication is 5’ to 3’

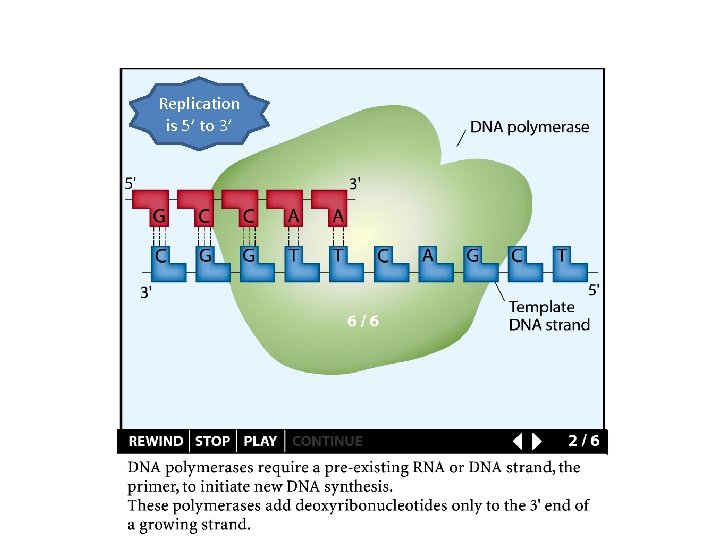
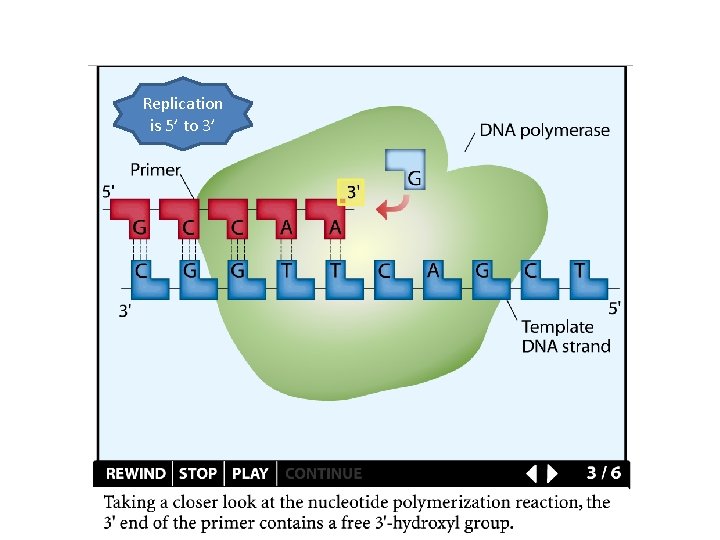
Replication is 5’ to 3’

Replication is 5’ to 3’

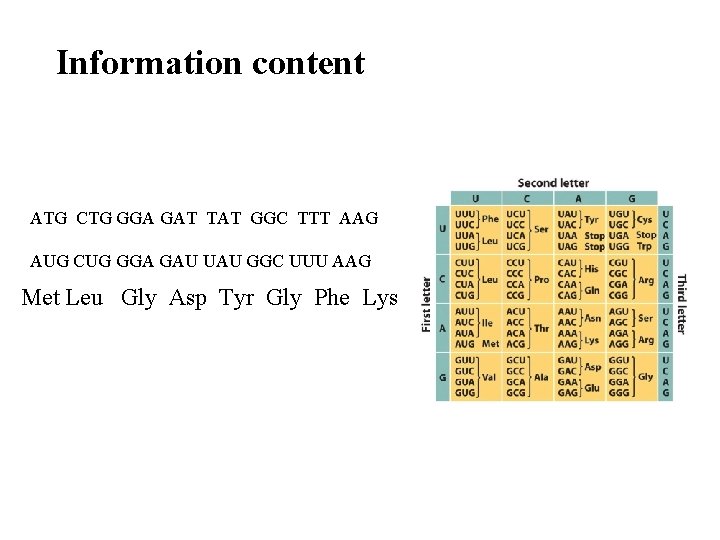
Information content ATG CTG GGA GAT TAT GGC TTT AAG AUG CUG GGA GAU UAU GGC UUU AAG Met Leu Gly Asp Tyr Gly Phe Lys

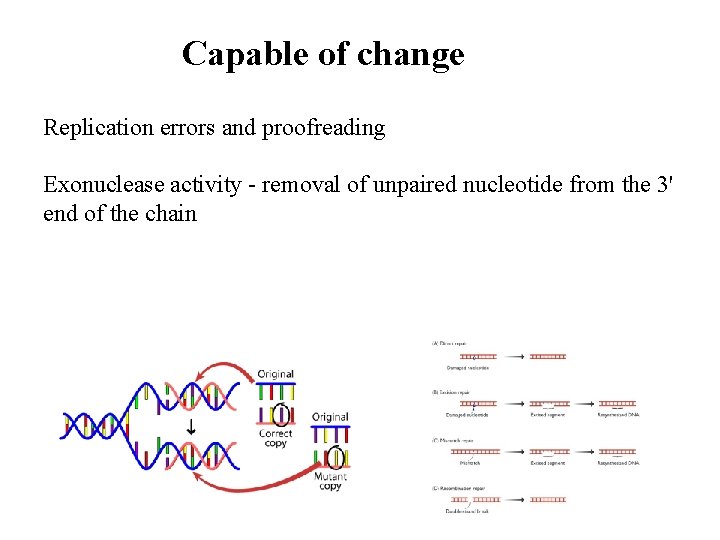
Capable of change Replication errors and proofreading Exonuclease activity - removal of unpaired nucleotide from the 3' end of the chain

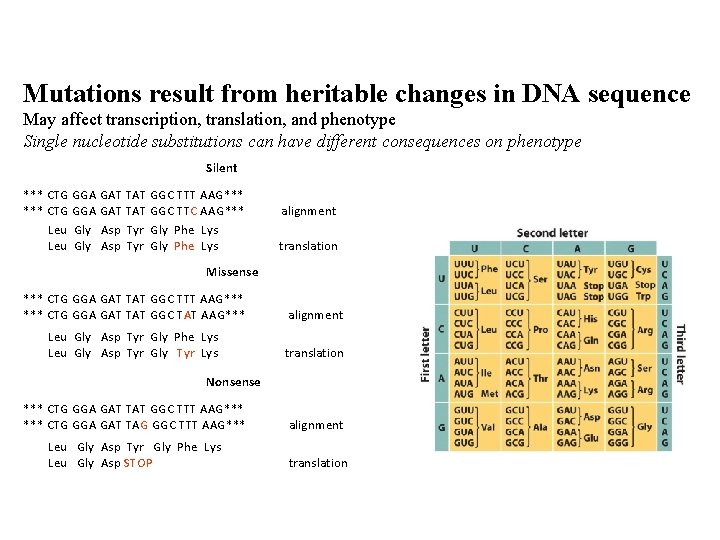
Mutations result from heritable changes in DNA sequence May affect transcription, translation, and phenotype Single nucleotide substitutions can have different consequences on phenotype Silent *** CTG GGA GAT TAT GGC TTT AAG*** CTG GGA GAT TAT GGC TTC AAG*** Leu Gly Asp Tyr Gly Phe Lys alignment translation Missense *** CTG GGA GAT TAT GGC TTT AAG*** CTG GGA GAT TAT GGC TAT AAG*** alignment Leu Gly Asp Tyr Gly Phe Lys Leu Gly Asp Tyr Gly Tyr Lys translation Nonsense *** CTG GGA GAT TAT GGC TTT AAG*** CTG GGA GAT TAG GGC TTT AAG*** alignment Leu Gly Asp Tyr Gly Phe Lys Leu Gly Asp STOP translation

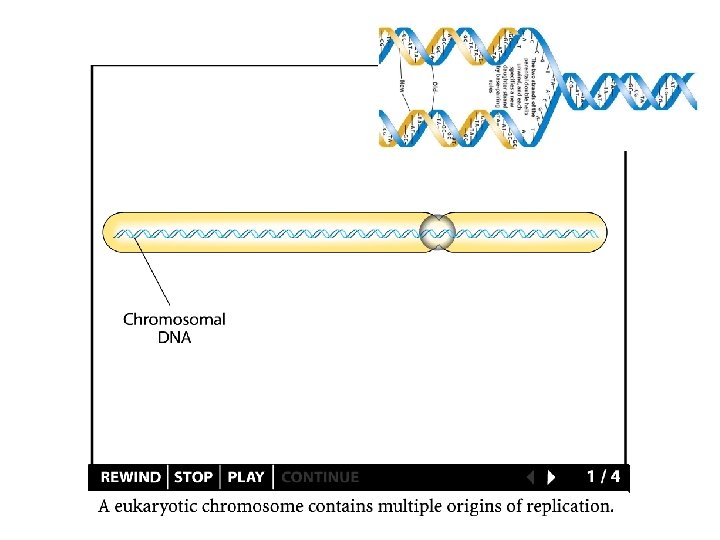
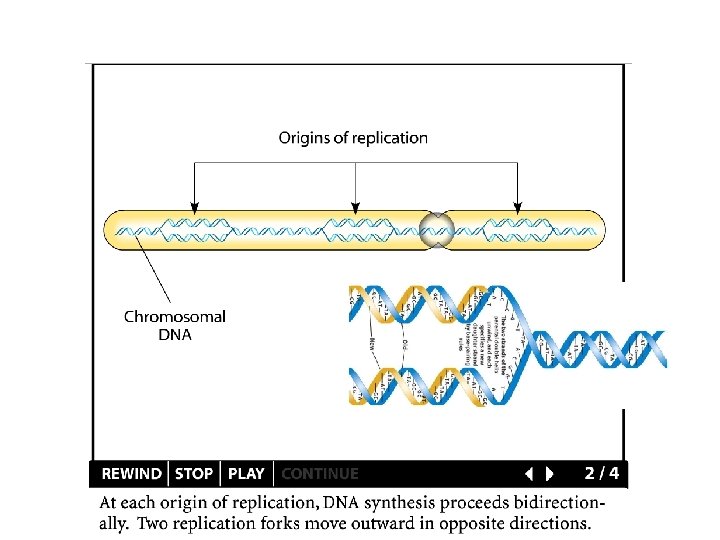
DNA Replication - in vivo Replication begins at an origin - proceeds bi-directionally In a higher plant chromosome - thousands of origins The size of the genome The rate of DNA replication The length of the S phase

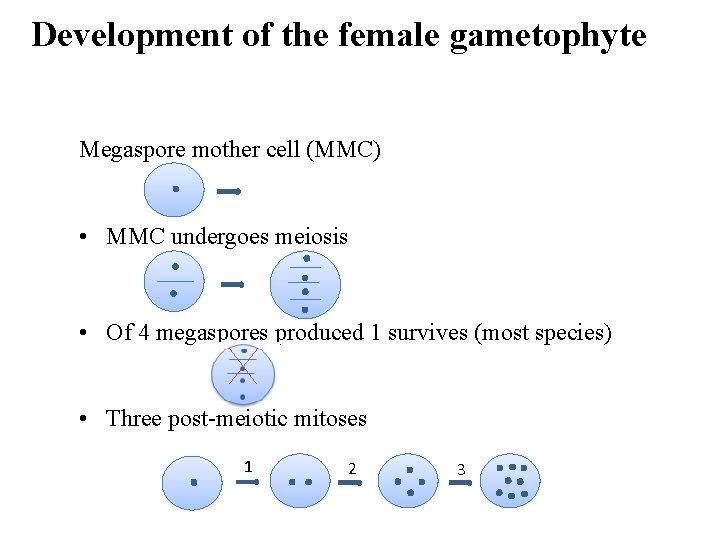
Development of the female gametophyte Megaspore mother cell (MMC) • MMC undergoes meiosis • Of 4 megaspores produced 1 survives (most species) • Three post-meiotic mitoses 1 2 3

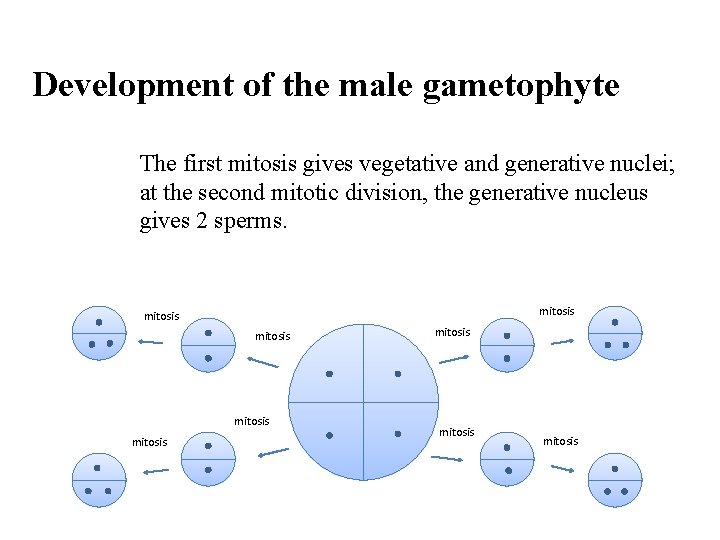
Development of the male gametophyte The first mitosis gives vegetative and generative nuclei; at the second mitotic division, the generative nucleus gives 2 sperms. mitosis mitosis





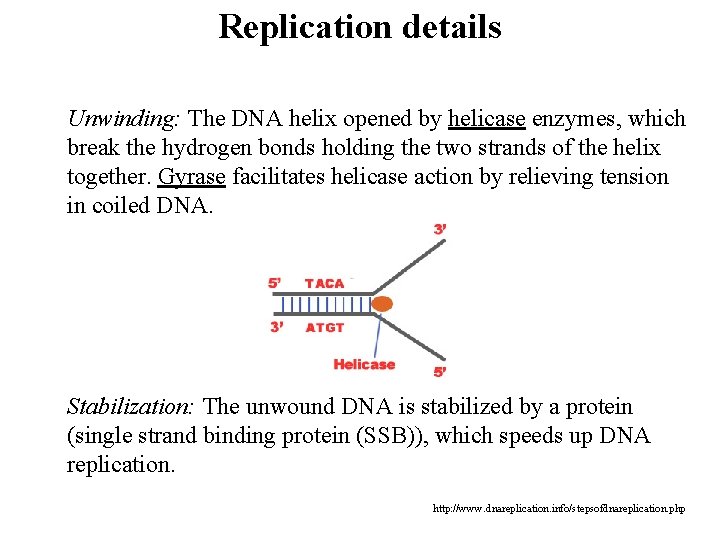
Replication details Unwinding: The DNA helix opened by helicase enzymes, which break the hydrogen bonds holding the two strands of the helix together. Gyrase facilitates helicase action by relieving tension in coiled DNA. Stabilization: The unwound DNA is stabilized by a protein (single strand binding protein (SSB)), which speeds up DNA replication. http: //www. dnareplication. info/stepsofdnareplication. php

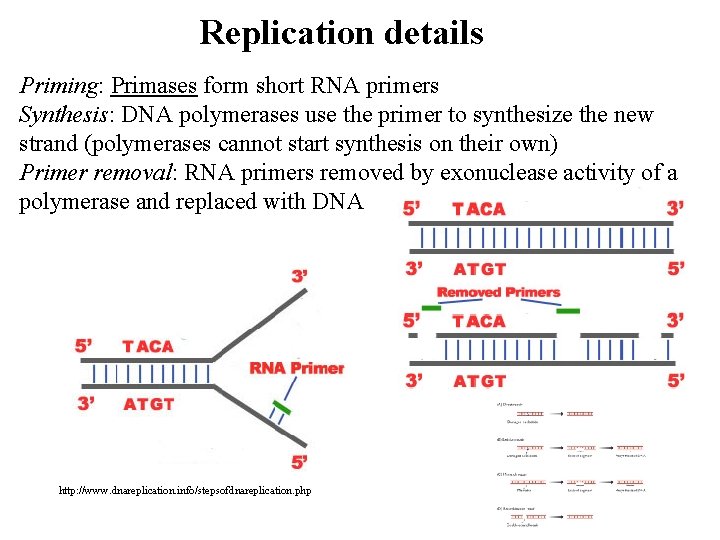
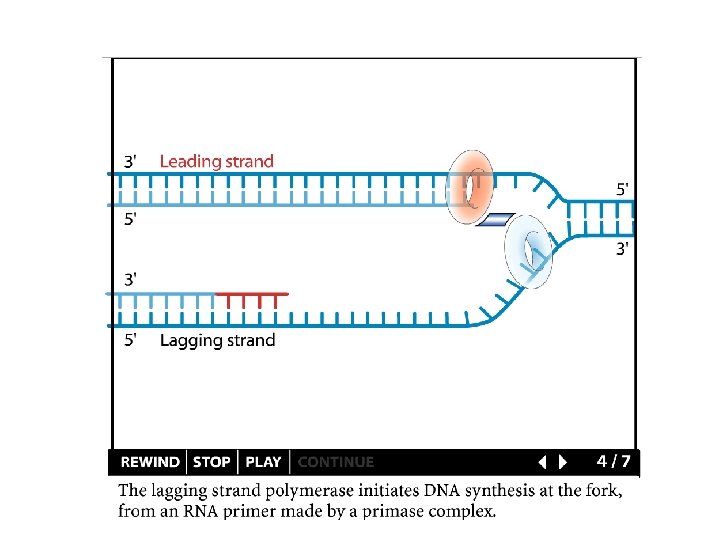
Replication details Priming: Primases form short RNA primers Synthesis: DNA polymerases use the primer to synthesize the new strand (polymerases cannot start synthesis on their own) Primer removal: RNA primers removed by exonuclease activity of a polymerase and replaced with DNA http: //www. dnareplication. info/stepsofdnareplication. php

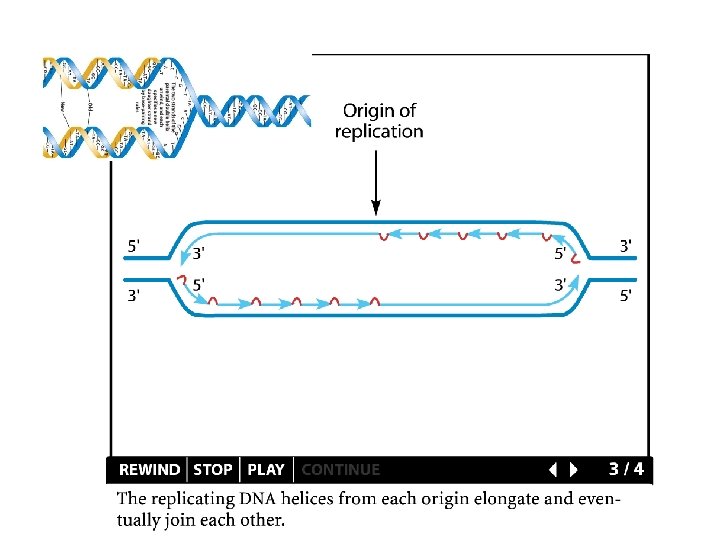
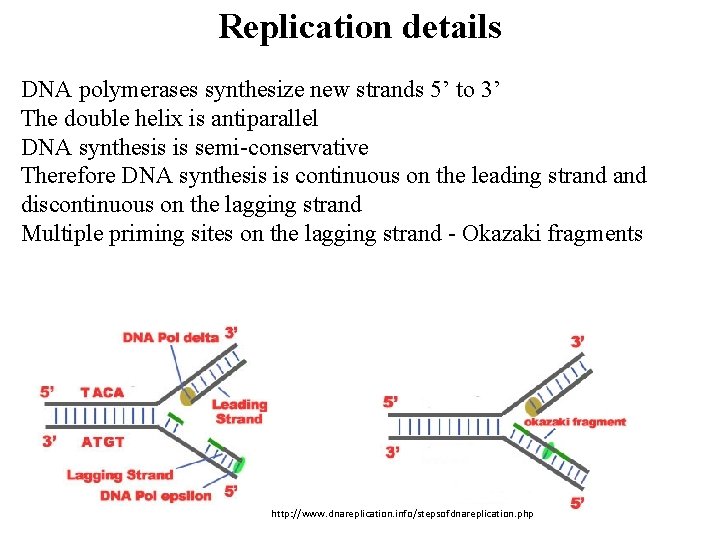
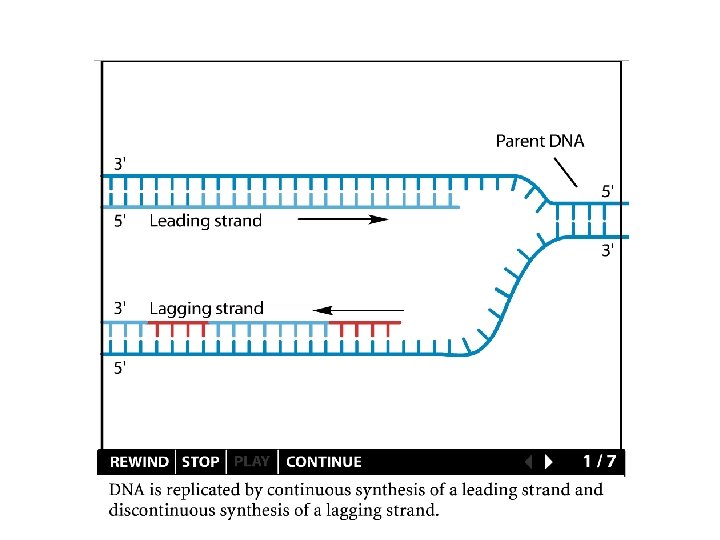
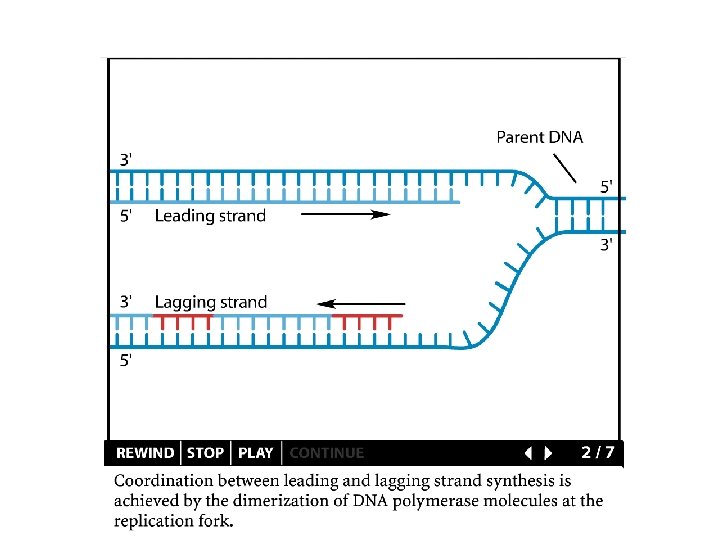
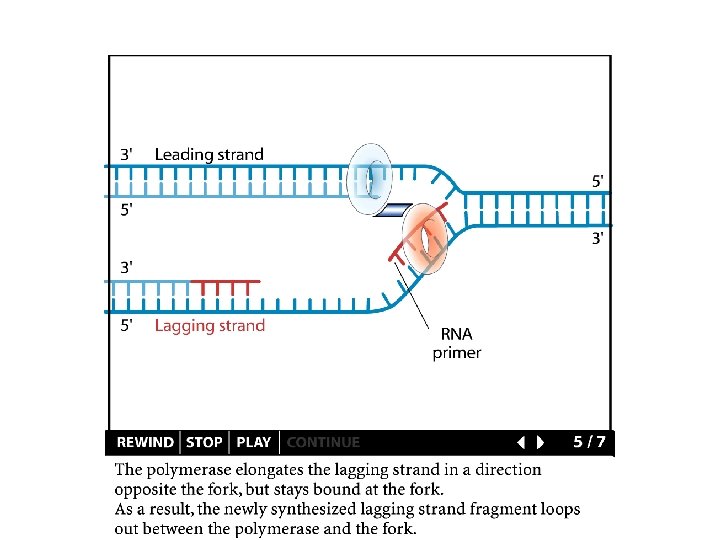
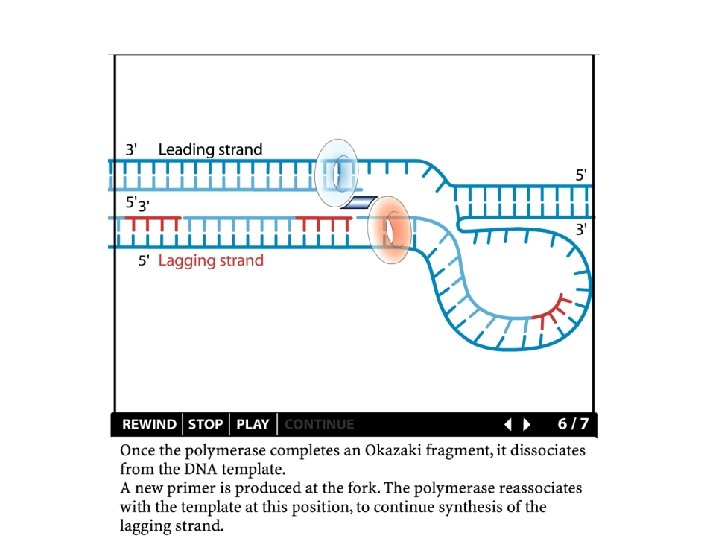
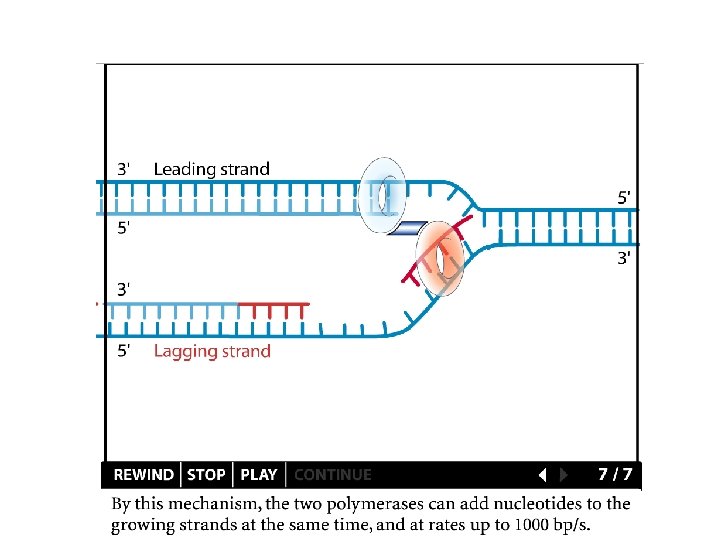
Replication details DNA polymerases synthesize new strands 5’ to 3’ The double helix is antiparallel DNA synthesis is semi-conservative Therefore DNA synthesis is continuous on the leading strand discontinuous on the lagging strand Multiple priming sites on the lagging strand - Okazaki fragments http: //www. dnareplication. info/stepsofdnareplication. php








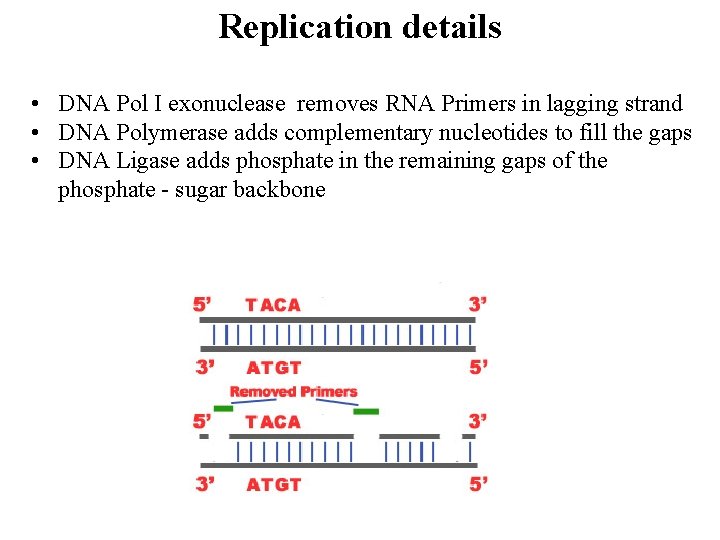
Replication details • DNA Pol I exonuclease removes RNA Primers in lagging strand • DNA Polymerase adds complementary nucleotides to fill the gaps • DNA Ligase adds phosphate in the remaining gaps of the phosphate - sugar backbone

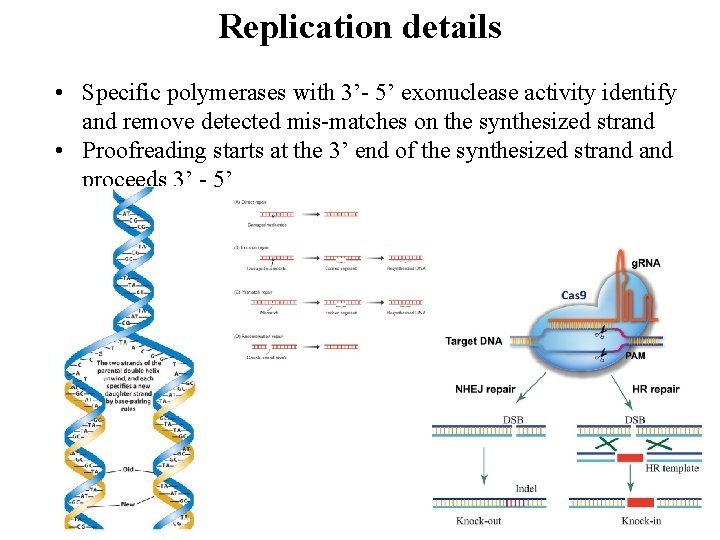
Replication details • Specific polymerases with 3’- 5’ exonuclease activity identify and remove detected mis-matches on the synthesized strand • Proofreading starts at the 3’ end of the synthesized strand proceeds 3’ - 5’

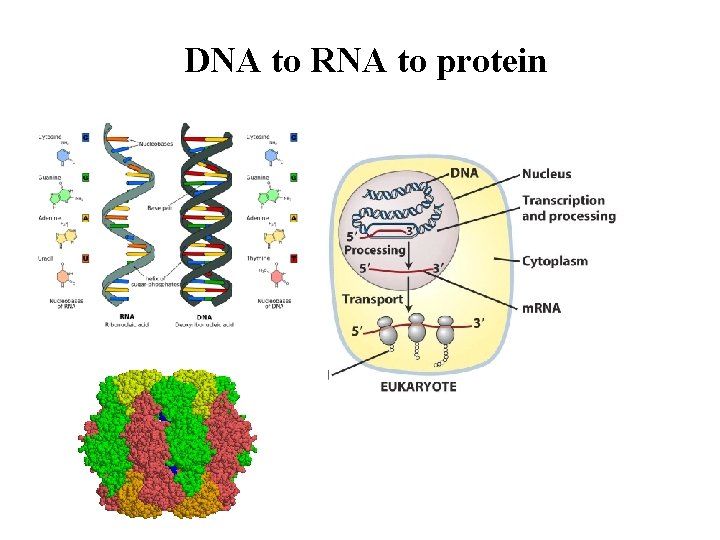
DNA to RNA to protein

From gene to phenotype “Central dogma” DNA → RNA → protein DNA Covered barley: Lemma and palea adhere to seed RNA Protein • • Other genes Pathways Environmental signals

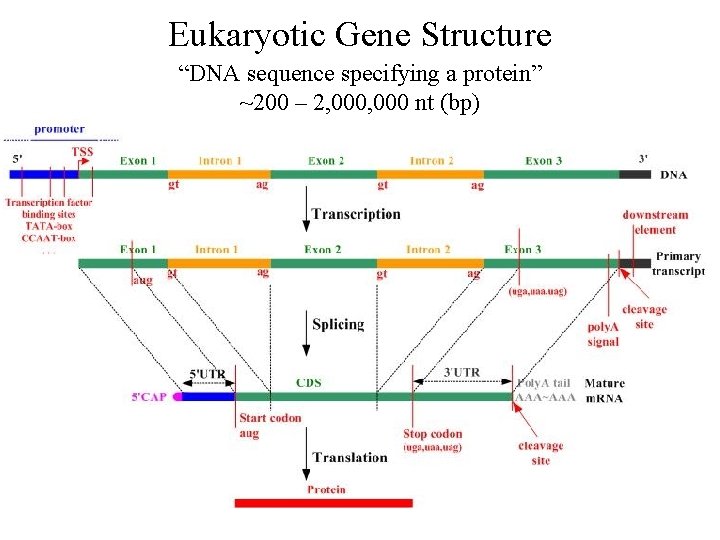
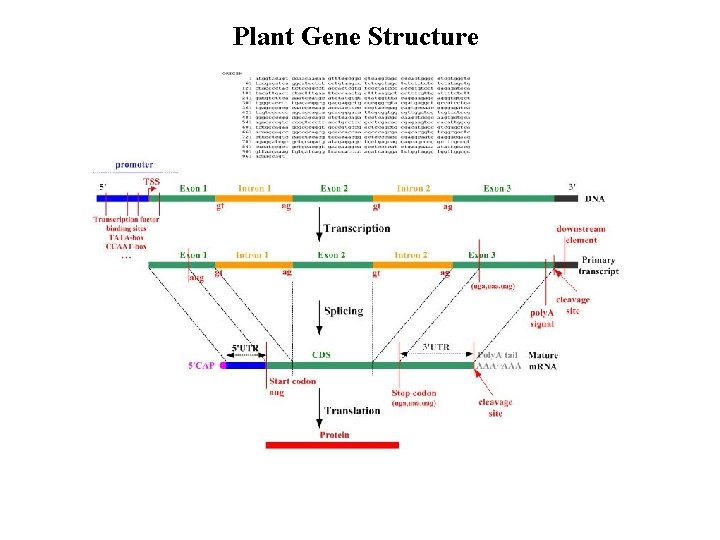
Eukaryotic Gene Structure “DNA sequence specifying a protein” ~200 – 2, 000 nt (bp)

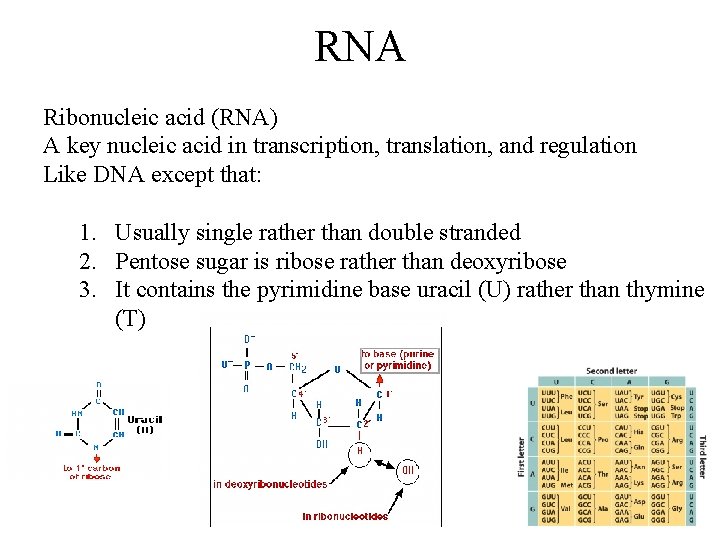
RNA Ribonucleic acid (RNA) A key nucleic acid in transcription, translation, and regulation Like DNA except that: 1. Usually single rather than double stranded 2. Pentose sugar is ribose rather than deoxyribose 3. It contains the pyrimidine base uracil (U) rather than thymine (T)

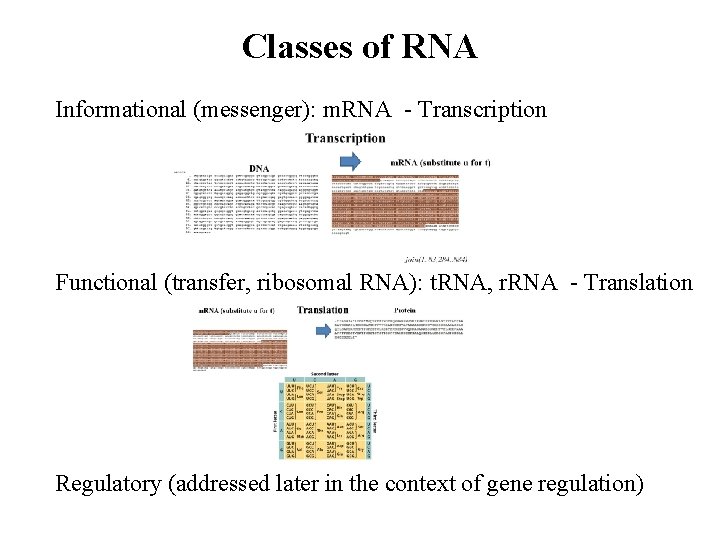
Classes of RNA Informational (messenger): m. RNA - Transcription Functional (transfer, ribosomal RNA): t. RNA, r. RNA - Translation Regulatory (addressed later in the context of gene regulation)

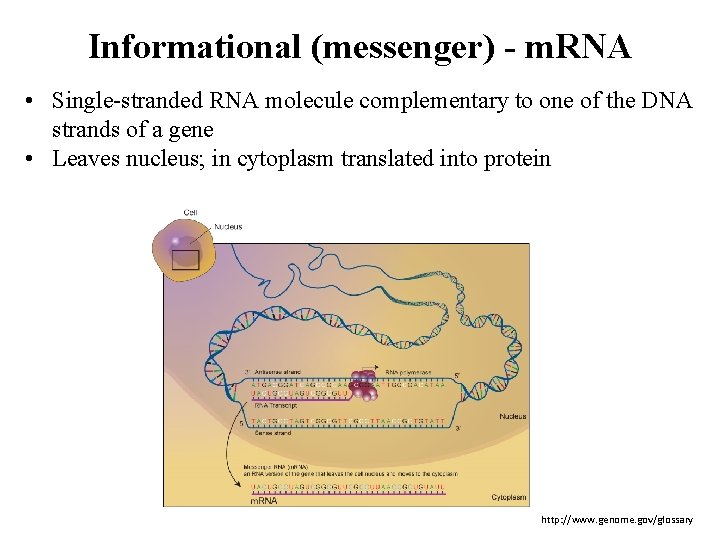
Informational (messenger) - m. RNA • Single-stranded RNA molecule complementary to one of the DNA strands of a gene • Leaves nucleus; in cytoplasm translated into protein http: //www. genome. gov/glossary

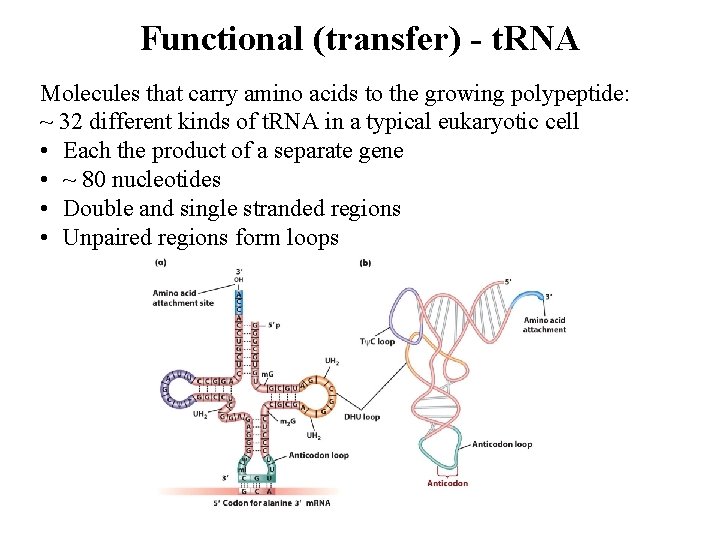
Functional (transfer) - t. RNA Molecules that carry amino acids to the growing polypeptide: ~ 32 different kinds of t. RNA in a typical eukaryotic cell • Each the product of a separate gene • ~ 80 nucleotides • Double and single stranded regions • Unpaired regions form loops

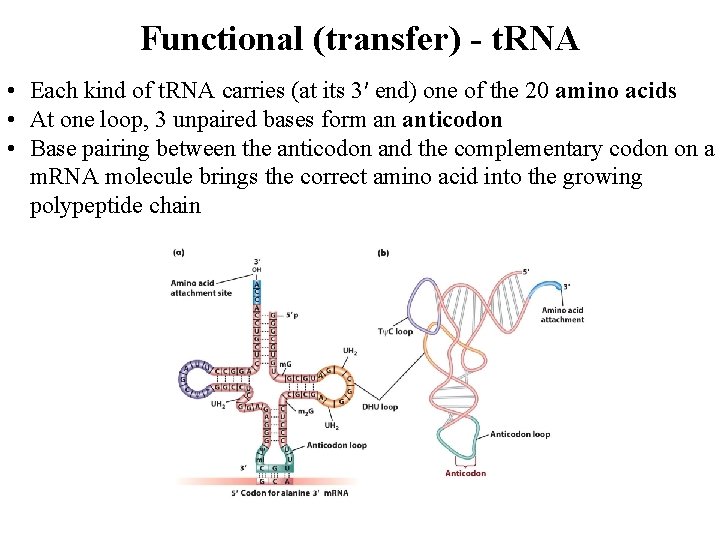
Functional (transfer) - t. RNA • Each kind of t. RNA carries (at its 3′ end) one of the 20 amino acids • At one loop, 3 unpaired bases form an anticodon • Base pairing between the anticodon and the complementary codon on a m. RNA molecule brings the correct amino acid into the growing polypeptide chain

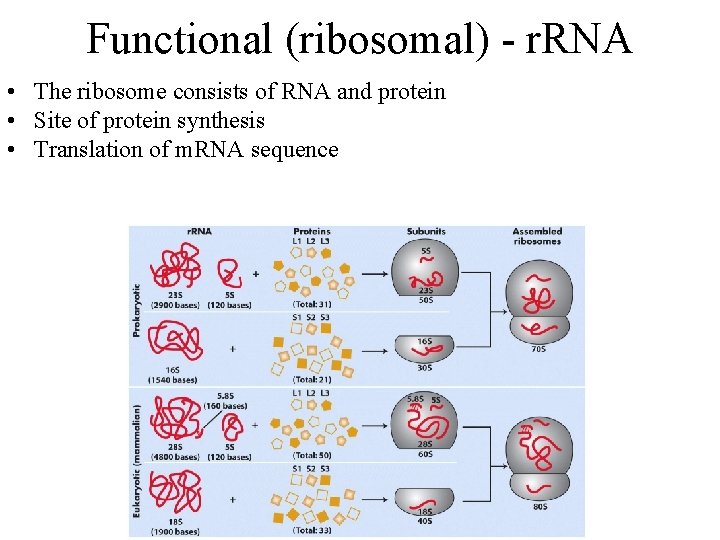
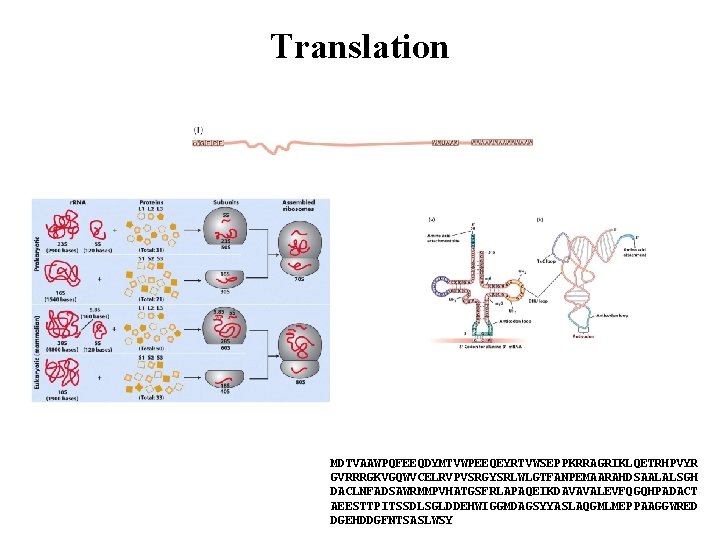
Functional (ribosomal) - r. RNA • The ribosome consists of RNA and protein • Site of protein synthesis • Translation of m. RNA sequence

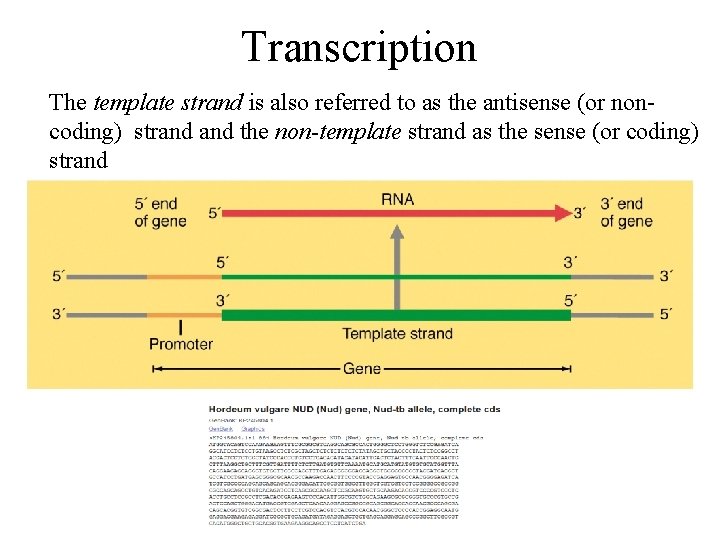
Transcription The template strand is also referred to as the antisense (or noncoding) strand the non-template strand as the sense (or coding) strand

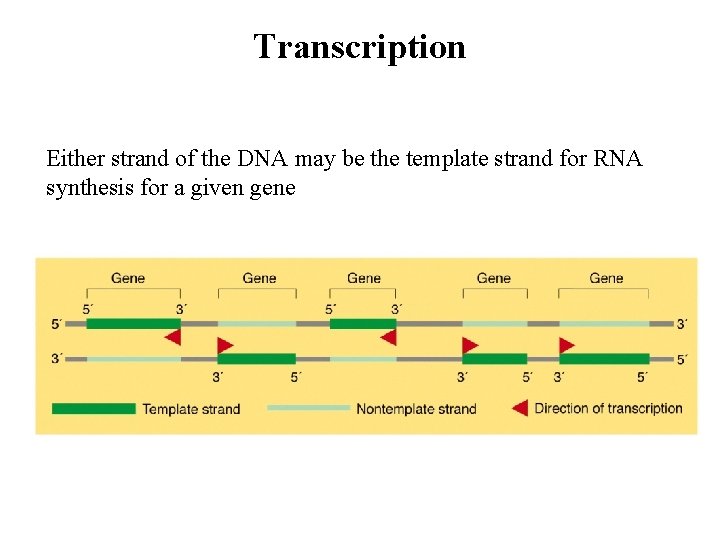
Transcription Either strand of the DNA may be the template strand for RNA synthesis for a given gene

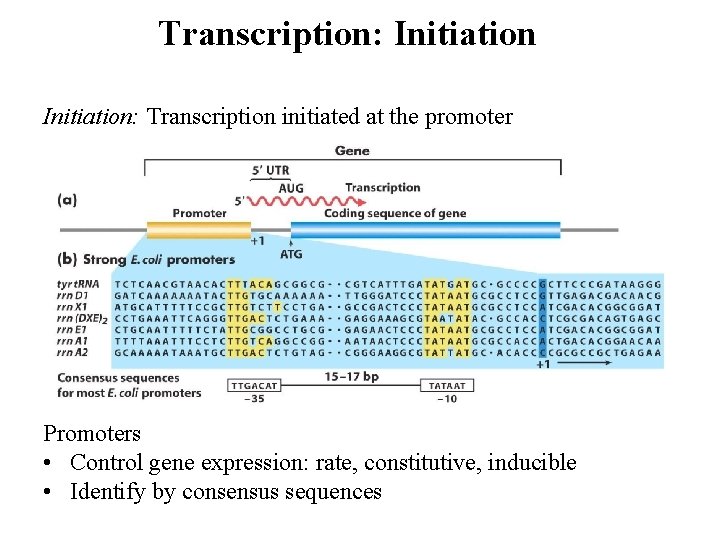
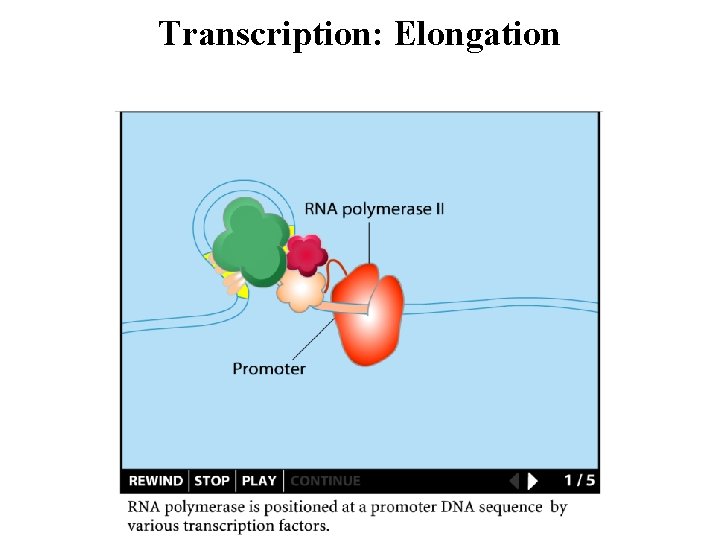
Transcription: Initiation: Transcription initiated at the promoter Promoters • Control gene expression: rate, constitutive, inducible • Identify by consensus sequences

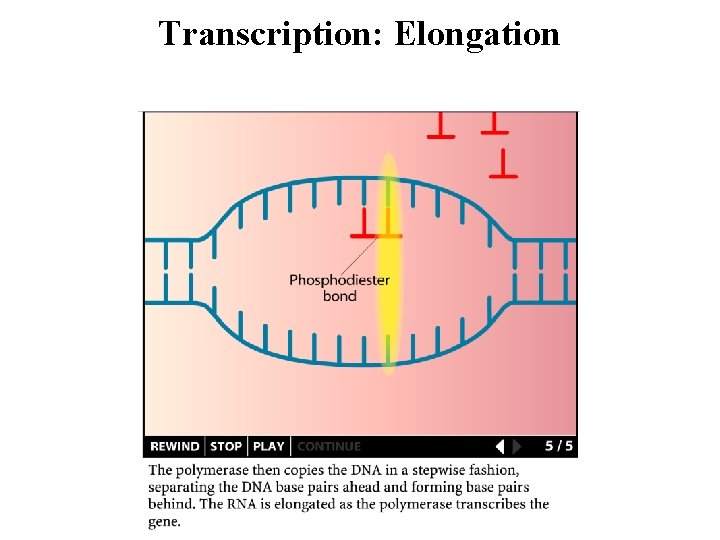
Transcription: Elongation

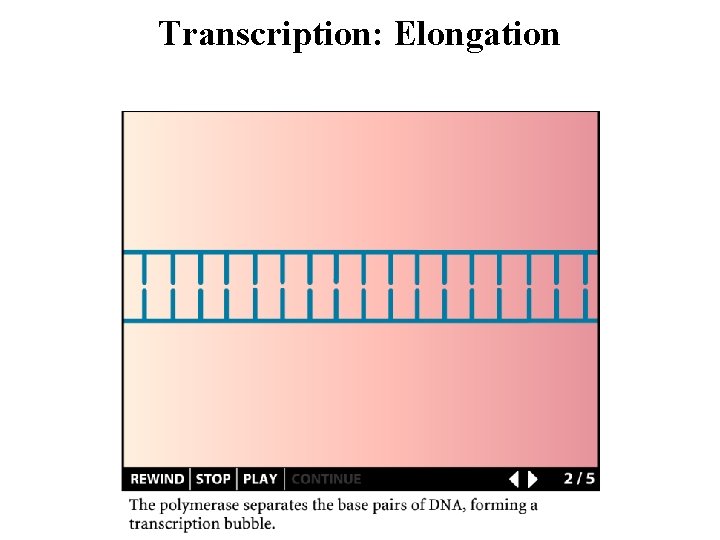
Transcription: Elongation

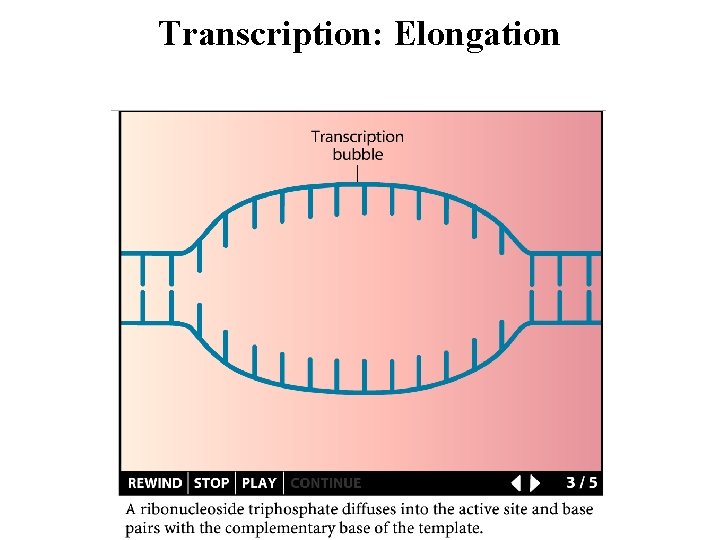
Transcription: Elongation

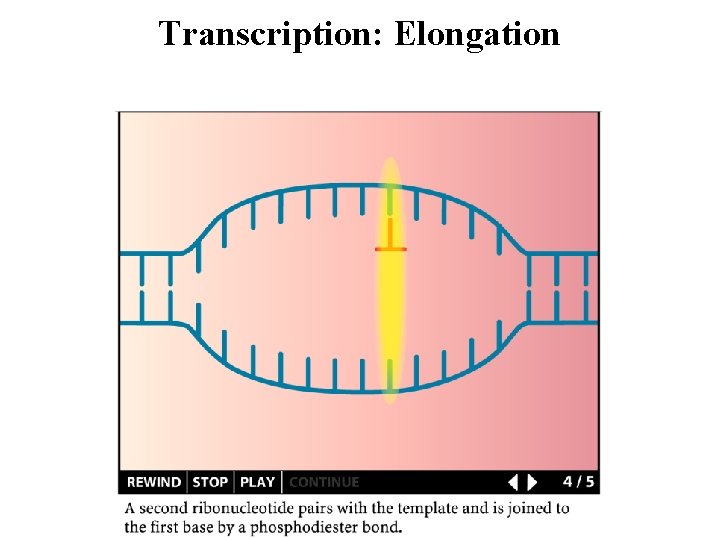
Transcription: Elongation

Transcription: Elongation

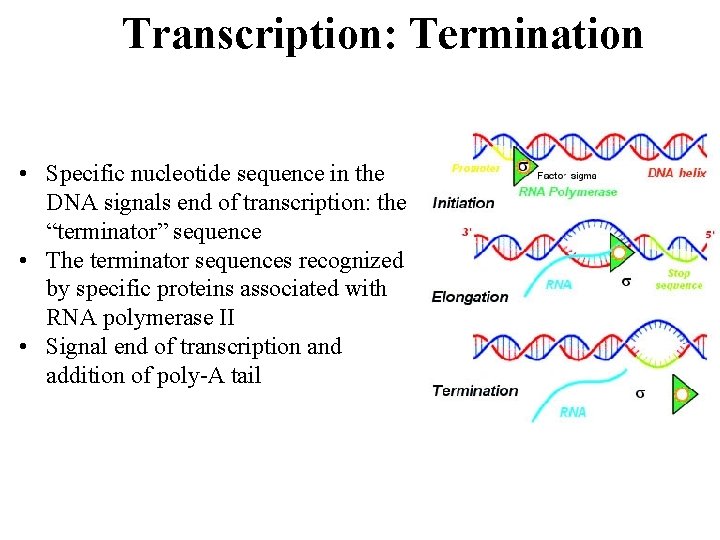
Transcription: Termination • Specific nucleotide sequence in the DNA signals end of transcription: the “terminator” sequence • The terminator sequences recognized by specific proteins associated with RNA polymerase II • Signal end of transcription and addition of poly-A tail

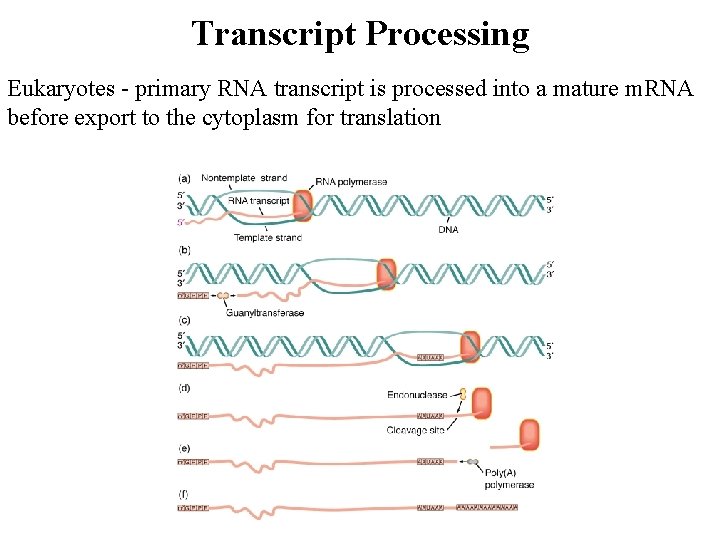
Transcript Processing Eukaryotes - primary RNA transcript is processed into a mature m. RNA before export to the cytoplasm for translation

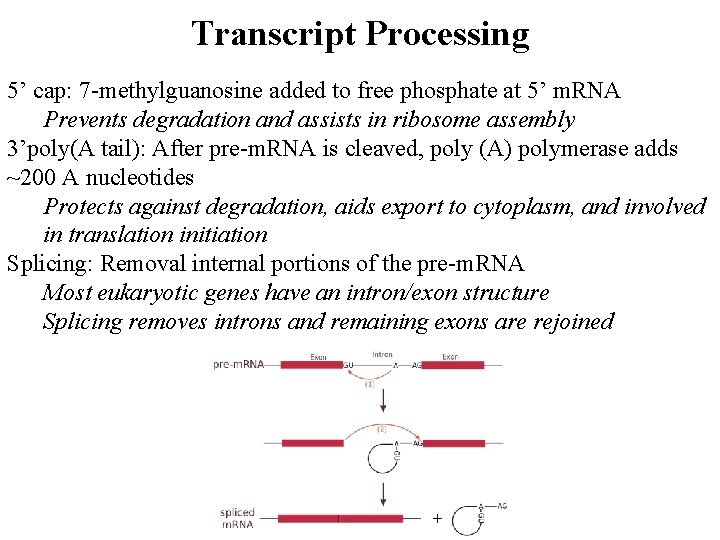
Transcript Processing 5’ cap: 7 -methylguanosine added to free phosphate at 5’ m. RNA Prevents degradation and assists in ribosome assembly 3’poly(A tail): After pre-m. RNA is cleaved, poly (A) polymerase adds ~200 A nucleotides Protects against degradation, aids export to cytoplasm, and involved in translation initiation Splicing: Removal internal portions of the pre-m. RNA Most eukaryotic genes have an intron/exon structure Splicing removes introns and remaining exons are rejoined

Plant Gene Structure

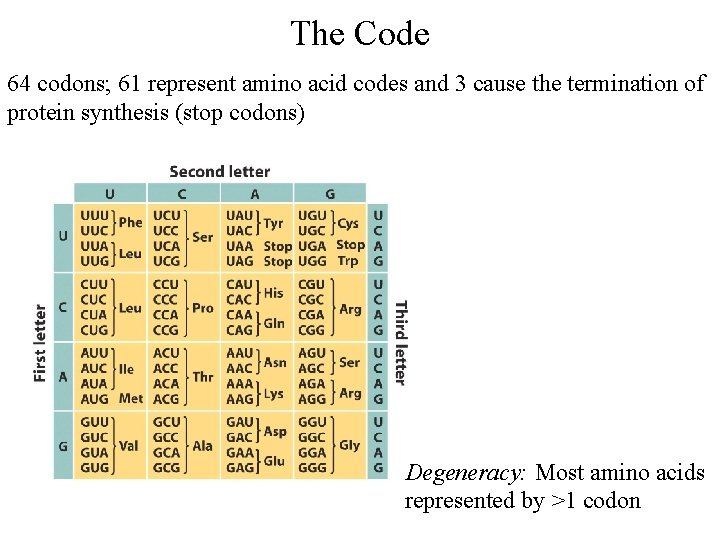
The Code 64 codons; 61 represent amino acid codes and 3 cause the termination of protein synthesis (stop codons) Degeneracy: Most amino acids represented by >1 codon

Translation MDTVAAWPQFEEQDYMTVWPEEQEYRTVWSEPPKRRAGRIKLQETRHPVYR GVRRRGKVGQWVCELRVPVSRGYSRLWLGTFANPEMAARAHDSAALALSGH DACLNFADSAWRMMPVHATGSFRLAPAQEIKDAVAVALEVFQGQHPADACT AEESTTPITSSDLSGLDDEHWIGGMDAGSYYASLAQGMLMEPPAAGGWRED DGEHDDGFNTSASLWSY

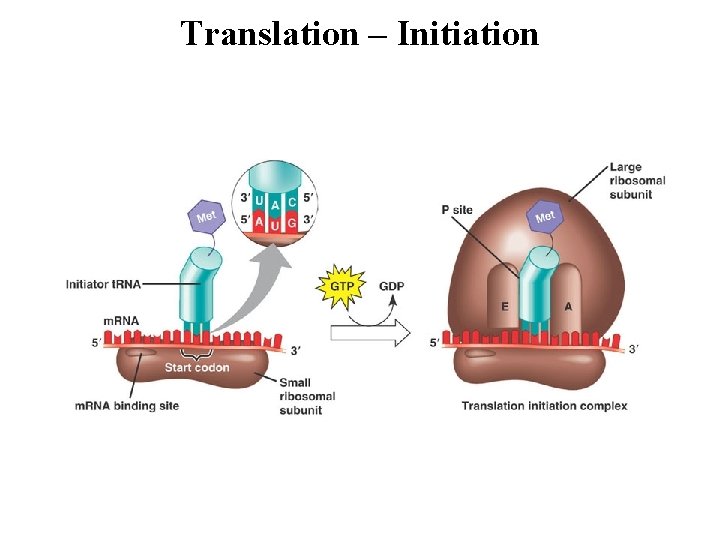
Translation – Initiation

Translation – Elongation

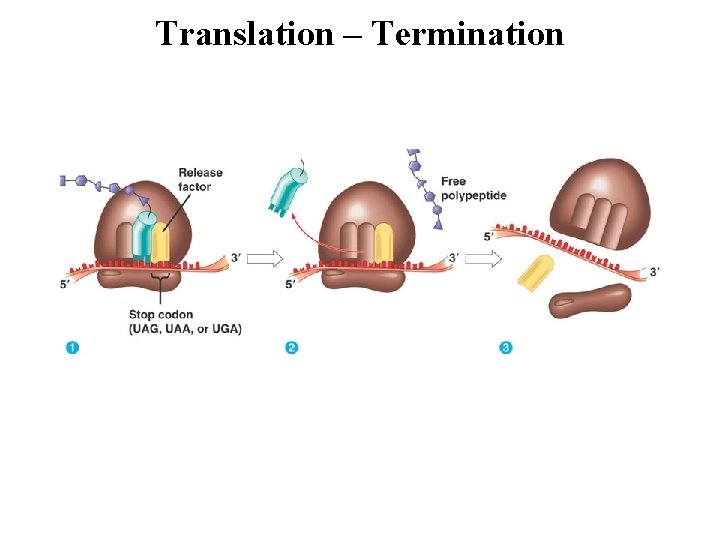
Translation – Termination

gene <1. . >77 /gene="CBF 2 A" m. RNA <1. . >772 /gene="CBF 2 A" /product="Hv. CBF 2 A” 5'UTR <1. . 12 /gene="CBF 2 A" CDS 13. . 678 /gene="CBF 2 A” /note="Hv. CBF 2 A-Dt; AP 2 domain CBF protein; putative CRT binding factor; monocot Hv. CBF 4 -subgroup member /codon_start=1 /product="Hv. CBF 2 A" /protein_id="AAX 23688. 1" /db_xref="GI: 60547429" /translation="MDTVAAWPQFEEQDYMTVWPEEQEYRTVWSEPPKRRAGRIKLQE TRHPVYRGVRRRGKVGQWVCELRVPVSRGYSRLWLGTFANPEMAARAHDSAALALSGH DACLNFADSAWRMMPVHATGSFRLAPAQEIKDAVAVALEVFQGQHPADACTAEESTTP ITSSDLSGLDDEHWIGGMDAGSYYASLAQGMLMEPPAAGGWREDDGEHDDGFNTSASL WSY" 3'UTR 679. . >772 /gene="CBF 2 A"

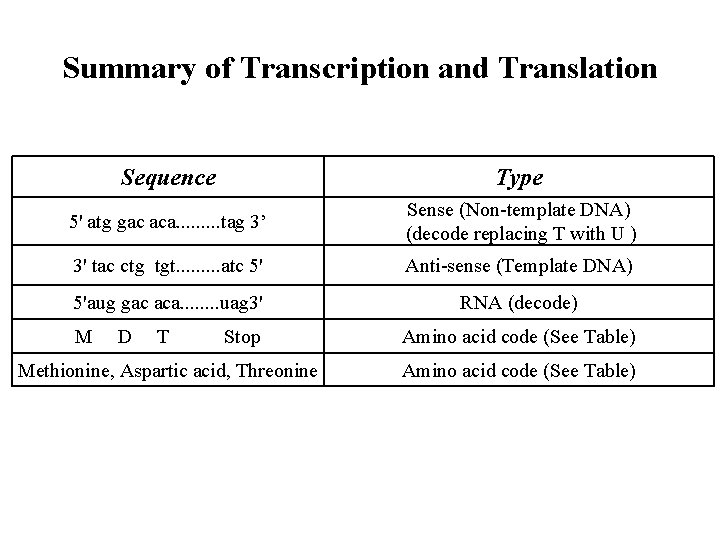
Summary of Transcription and Translation Sequence Type 5' atg gac aca. . tag 3’ Sense (Non-template DNA) (decode replacing T with U ) 3' tac ctg tgt. . atc 5' Anti-sense (Template DNA) 5'aug gac aca. . . . uag 3' RNA (decode) M D T Stop Amino acid code (See Table) Methionine, Aspartic acid, Threonine Amino acid code (See Table)

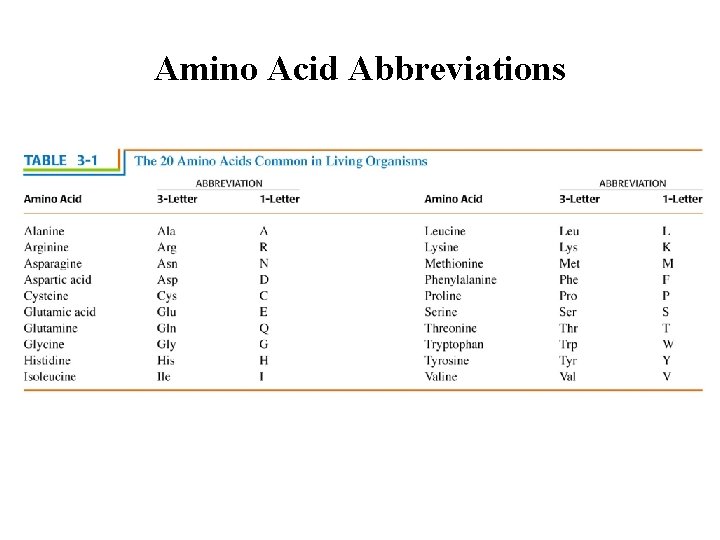
Amino Acid Abbreviations

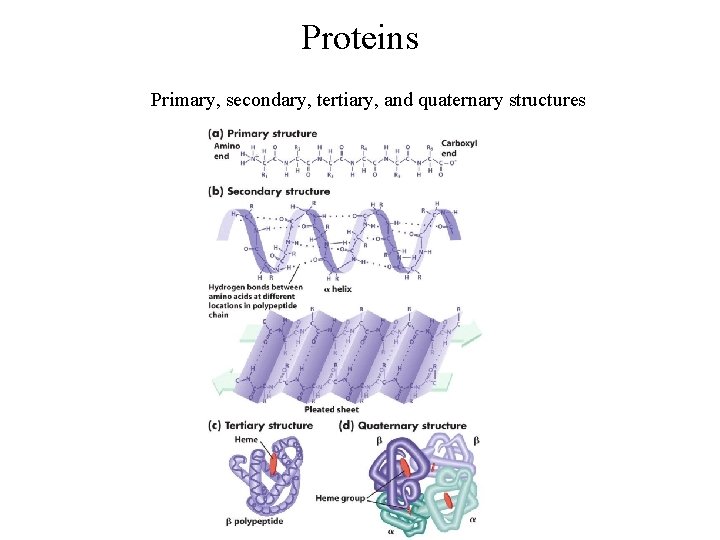
Proteins Primary, secondary, tertiary, and quaternary structures

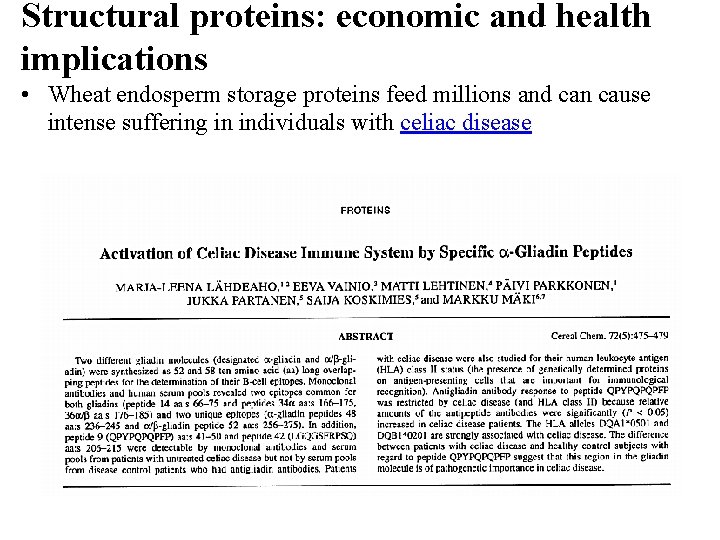
Structural proteins: economic and health implications • Wheat endosperm storage proteins feed millions and can cause intense suffering in individuals with celiac disease

Transcription, Translation, Phenotype Fragrance in rice