From Asymmetric Exclusion Processes to Protein Synthesis Beate

From Asymmetric Exclusion Processes to Protein Synthesis Beate Schmittmann Physics Department, Virginia Tech with Jiajia Dong (Hamline U. ) and Royce Zia (Virginia Tech), and many thanks to Leah Shaw (William & Mary). Workshop on Nonequilibrium dynamics of spatially extended interacting particle systems January 11 -13, 2010 Funded by the Division of Materials Research, NSF

Outline: • Basic facts about protein synthesis • A simple model: TASEP with locally varying rates – Currents and density profiles for one and two slow codons – “point” particles – “extended” objects – Real genes • Conclusions and open questions

Protein synthesis Two steps: • Transcription: DNA RNA • Translation: RNA Protein Image courtesy of National Health Museum
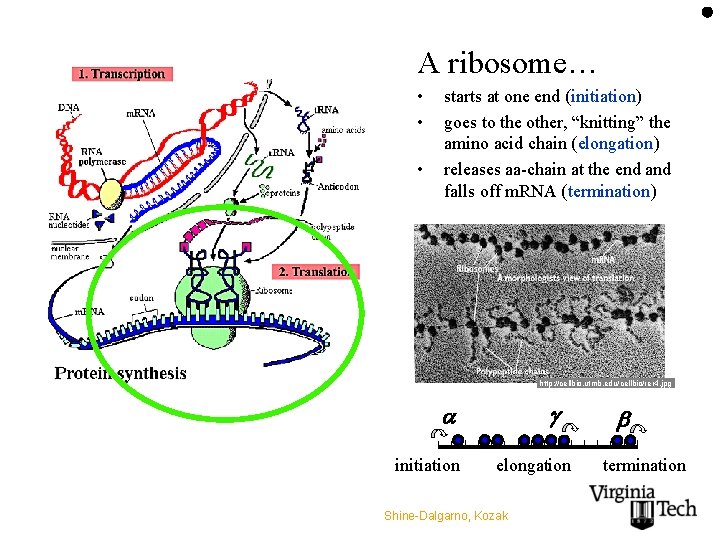
A ribosome… • • • starts at one end (initiation) goes to the other, “knitting” the amino acid chain (elongation) releases aa-chain at the end and falls off m. RNA (termination) Before one falls off, another one starts! http: //cellbio. utmb. edu/cellbio/rer 4. jpg initiation elongation Shine-Dalgarno, Kozak termination

Knitting the aa into the polypeptide chain Left: http: //www. emc. maricopa. edu/faculty/farabee/BIOBK/Bio. Bookgloss. E. html Right: cellbio. utmb. edu/cellbio/ribosome. htm; also Alberts et al, 1994

Some interesting features: • In E. coli, 61 codons code for 20 amino acids, mediated by 46 t. RNAs Synonymous codons code for same amino acid; Degeneracy ranges from 1 to 6 • t. RNA concentrations can vary by orders of magnitude • Translation rate believed to be determined by t. RNA concentrations “Fast” and “slow” codons

Example: Leucine in E. Coli t. RNA codon H. Dong, L. Nilsson, and C. G. Kurland, J. Mol. Biol. 1996

Some interesting features: • In E. coli, 61 codons code for 20 amino acids, mediated by 46 t. RNAs Synonymous codons code for same amino acid; Degeneracy ranges from 1 to 6 • t. RNA concentrations can vary by orders of magnitude • Translation rate believed to be determined by t. RNA concentrations “Fast” and “slow” codons • Codon bias: In highly expressed genes, “fast” codons appear more frequently than their “slower” synonymous counterparts

Towards a theoretical description: • Translation is a one-dimensional, unidirectional process with excluded volume interactions • Suggests modeling via a totally asymmetric exclusion process

Totally asymmetric simple exclusion process The model: TASEP of point particles • Open chain: – sites are occupied or empty – particles hop with rate 1 to empty nearest-neighbor sites on the right – particles hop on (off) the chain with rate ( ) – random sequential dynamics (easily simulated!) …… • Ring: much simpler The proto model: F. Spitzer, Adv. Math. 5, 246 (1970)

Why study TASEP ? • Mathematicians: “Consider… this stochastic process” • Biologists: simple minded model for protein synthesis • Physicists: – Non-equilibrium statistical mechanics – Interacting systems with dynamics that violate detailed balance, time reversal – Novel states and stationary distributions – Many other potential applications
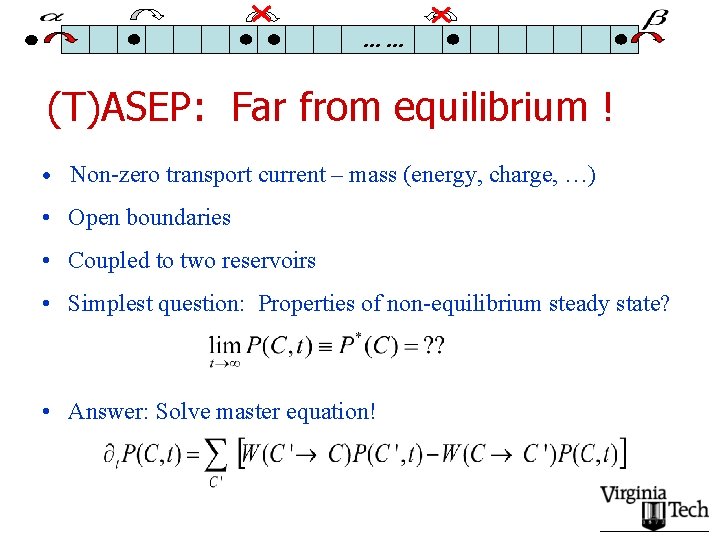
…… (T)ASEP: Far from equilibrium ! • Non-zero transport current – mass (energy, charge, …) • Open boundaries • Coupled to two reservoirs • Simplest question: Properties of non-equilibrium steady state? • Answer: Solve master equation!
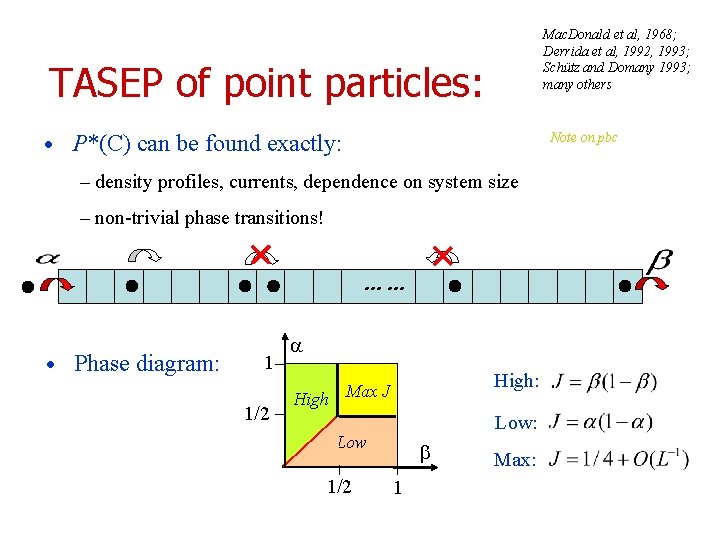
Mac. Donald et al, 1968; Derrida et al, 1992, 1993; Schütz and Domany 1993; many others TASEP of point particles: • P*(C) can be found exactly: Note on pbc – density profiles, currents, dependence on system size – non-trivial phase transitions! …… • Phase diagram: 1 1/2 High: High Max J Low: Low 1/2 1 Max:

Towards a theoretical description: • Translation is a one-dimensional, unidirectional process with excluded volume interactions • Suggests modeling via a totally asymmetric exclusion process • Modifications: – Translation rates are spatially non-uniform; start with one or two slow codons, (A. Kolomeisky, 1998; Chou & Lakatos, 2004) then consider a whole gene – Ribosomes are extended objects (cover about 10 – 12 codons); start with pointlike objects, then consider different sizes (L. B. Shaw et al, 2003, 2004) • Goal: Explore the effect of “bottle necks” (rates, location) and xxxribosome size

TASEP with bottle necks: • To model the effects of one or two slow codons: – change hopping rates locally to q 1 – for simplicity, choose = = 1 q q …… x y • Measure current ( protein production rate) and density profile: – as a function of x, y and q
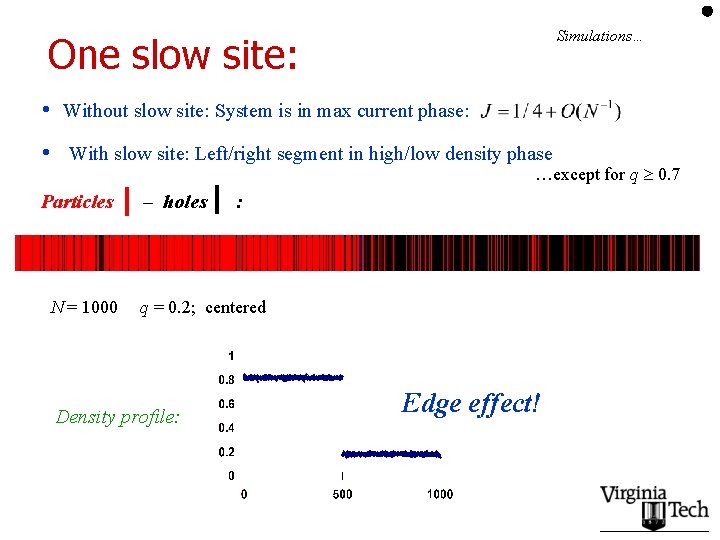
Simulations… One slow site: • Without slow site: System is in max current phase: • With slow site: Left/right segment in high/low density phase …except for q 0. 7 Particles N = 1000 – holes : q = 0. 2; centered Density profile: Edge effect!

Simulations… Edge effect: N = 1000, q = 0. 6 Current: Density profiles: site Mean-field theory: A. Kolomeisky, 1998 Maximized at q=0. 49: 2. 5% k=1: good results from FSMFT

Simulations… Two slow sites: … and extension of MFT Particles – holes: L = 1000; q 1 = q 2 = 0. 2; separated by 500 sites Typical density profiles: q 1 = q 2 = 0. 2 q 1 = q 2 = 0. 6
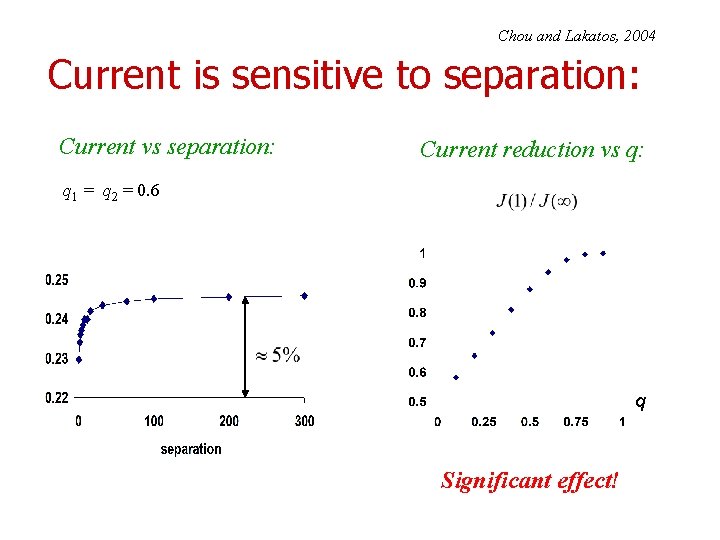
Chou and Lakatos, 2004 Current is sensitive to separation: Current vs separation: Current reduction vs q: q 1 = q 2 = 0. 6 q Significant effect!

Chou and Lakatos, PLR 2004; Dong, Schmittmann, Zia JSP 2007 First set of conclusions: • To maximize current, i. e. , protein synthesis rate: – Slow codons should be spaced as far apart as possible! • Check effect of particle size! Note: • Two slow sites with q 1 q 2 : Slowest site determines current • Fast site(s) : Significant effects on profiles; none on currents

Effect of particle size, l …… • Entry: – only if first l sites are free; then, whole particle enters with rate • Hopping: – left-most site is “reader”, determines local rate • Exit: – hops out gradually, “reader” leaves with rate β Lakatos and Chou, JPA 36, 2027 (2003): Complete entry and incremental exit

Phase diagram: 1 Mc. Donald and Gibbs, 1969; Lakatos and Chou, 2003; Shaw et al. , 2003 Results based on mean-field analysis or extremal principle; no longer exact but in good agreement with simulations. Max J High Low 1 • High: • Low: • Max:

Simulations… One slow site: N = 1000, q = 0. 2, x = 82 • Without slow site: System is in max current phase. • With slow site: Left/right segment in high/low density phase l = 01 l = 06 l = 12 Edge effect! Long tails! Coverage density profile (all occupied sites) Reader density profile (only sites occupied by readers)
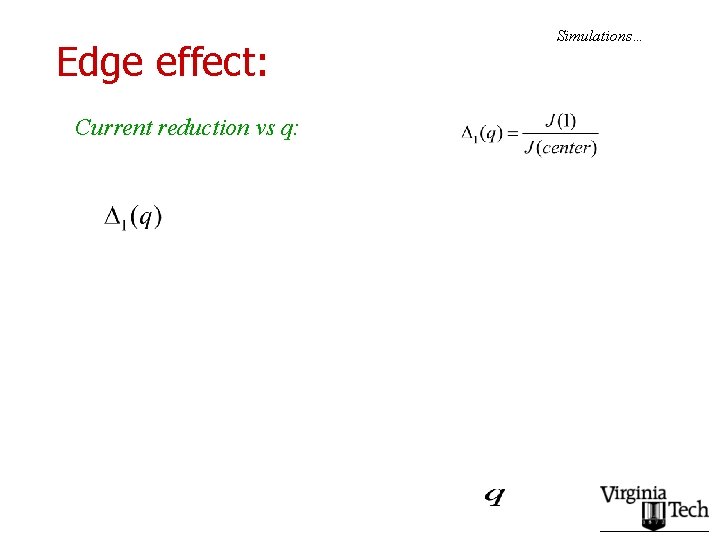
Edge effect: Current reduction vs q: Simulations…

Two slow sites: Shock still develops! Coverage density profile: Simulations… N = 1000, q = 0. 2 l = 01 l = 02 l = 06 l = 12 Reader density profile:
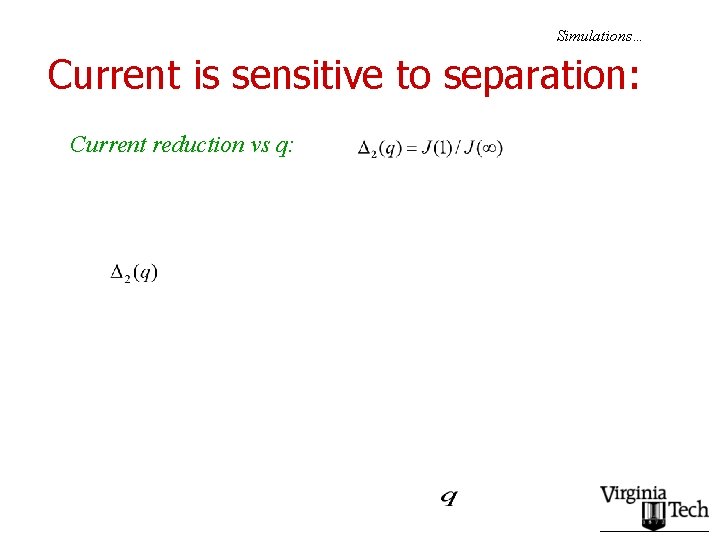
Simulations… Current is sensitive to separation: Current reduction vs q:

Second set of conclusions: • The basic conclusion of the point particle study remains valid: – Currents are maximized if slow codons are spaced as far apart as possible. – Edge effect becomes more dramatic, as l increases • Real genes?
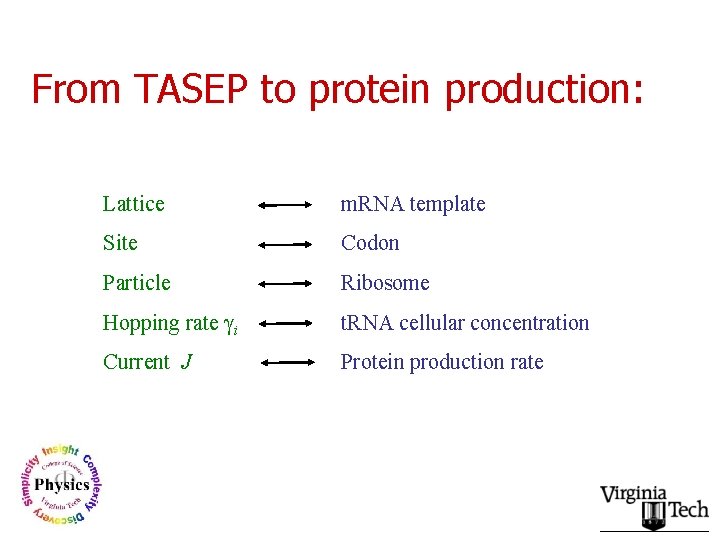
From TASEP to protein production: Lattice m. RNA template Site Codon Particle Ribosome Hopping rate γi t. RNA cellular concentration Current J Protein production rate
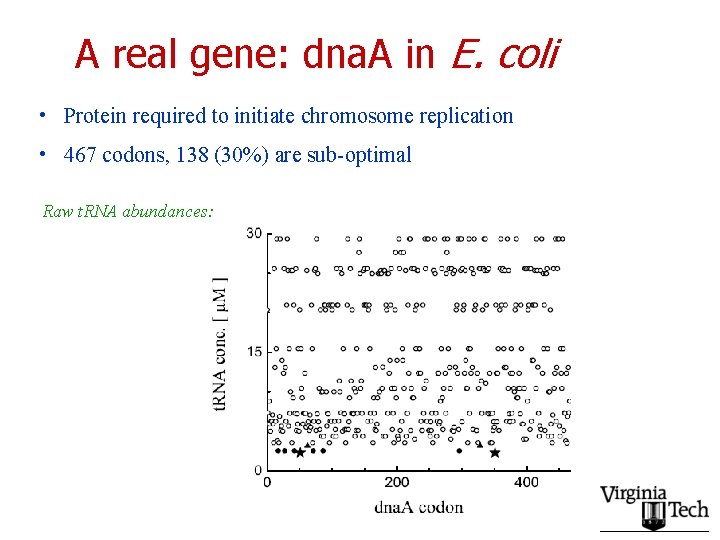
A real gene: dna. A in E. coli • Protein required to initiate chromosome replication • 467 codons, 138 (30%) are sub-optimal Raw t. RNA abundances:

Optimize: J ΔJ original (wild) optimal abysmal 0. 011455 0. 017514 0. 007115 + 53 % 38 % (138 replacements) (225 replacements) highest wild ~ 1. 5 ~ wild lowest
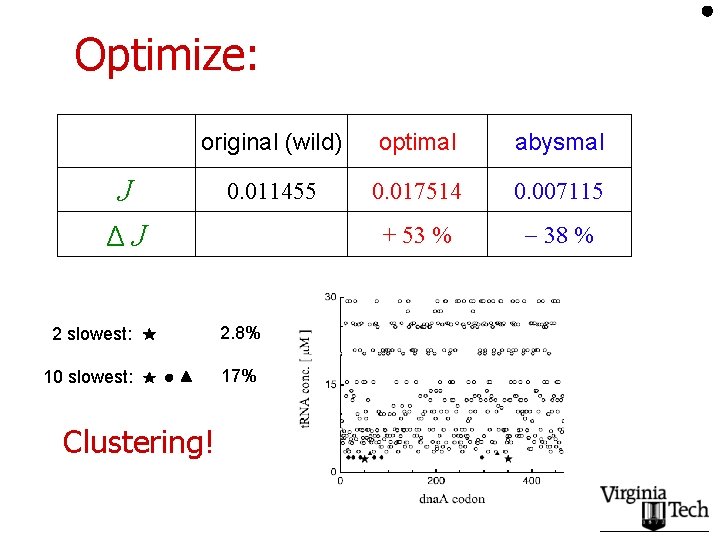
Optimize: original (wild) optimal abysmal 0. 011455 0. 017514 0. 007115 + 53 % 38 % J ΔJ 2 slowest: 2. 8% 10 slowest: 17% Clustering!

Clustering is important: • Introduce “coarse-grained” rate: • K 1 is time needed to traverse l consecutive sites Shaw, Zia, and Lee PRE 2003
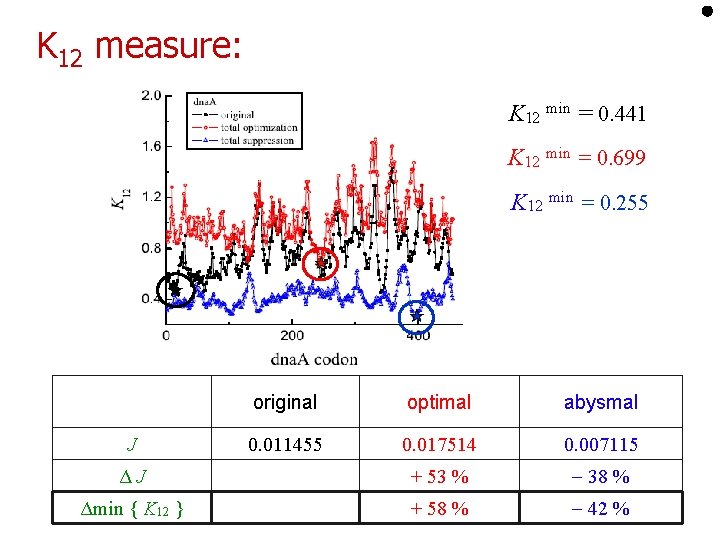
K 12 measure: K 12 min = 0. 441 K 12 min = 0. 699 K 12 min = 0. 255 original optimal abysmal 0. 011455 0. 017514 0. 007115 ΔJ + 53 % 38 % Δmin { K 12 } + 58 % 42 % J

Several sequences – same protein:
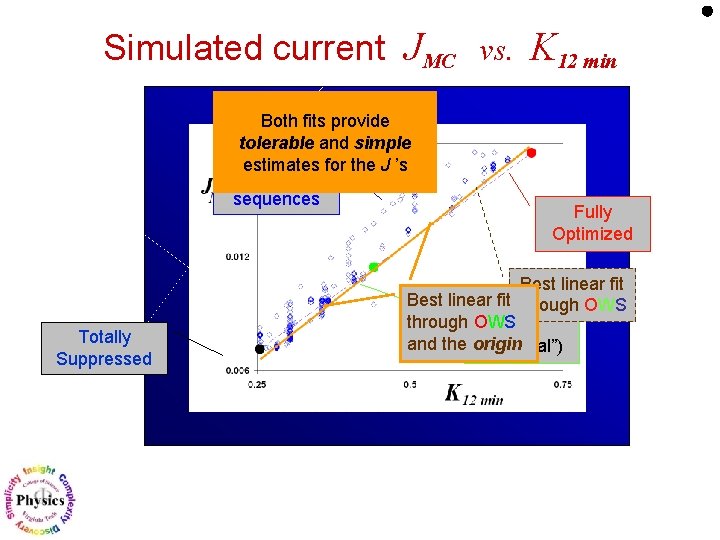
Simulated current JMC vs. K 12 min Both fits provide tolerable and simple estimates for the J ’s 700 other sequences Totally Suppressed Fully Optimized Best linear fit through OWS Wild and the origin (“original”)

Simulated current JMC vs. K 12 min Similar results for 10 other genes in E. coli Example of lac. I : (with just 5 other randomly generated sequences) Slopes are ~10% of each other. J ~ const. K 12 min DNA-binding transcriptional repressor ? ? ?
Conclusions: • Protein production can be increased significantly by a few xxtargeted removals of bottlenecks and clustered bottlenecks. • K measure provides simple estimate of changes in production rates • Extensions: Initiation-rate limited m. RNA; finite ribosome xxsupply; polycistronic m. RNA; parallel translation of multiple xxm. RNAs; and many other issues. • Experiments! J. J. Dong, B. Schmittmann, and R. K. P. Zia, J. Stat. Phys. 128, 21 (2007); Phys. Rev. E 76, 051113 (2007); J. Phys. A 42, 015002 (2009) J. J. Dong, Ph. D thesis. Virginia Tech (May 2008)
- Slides: 37