Frederick Griffith 1928 Conclusion living R bacteria transformed
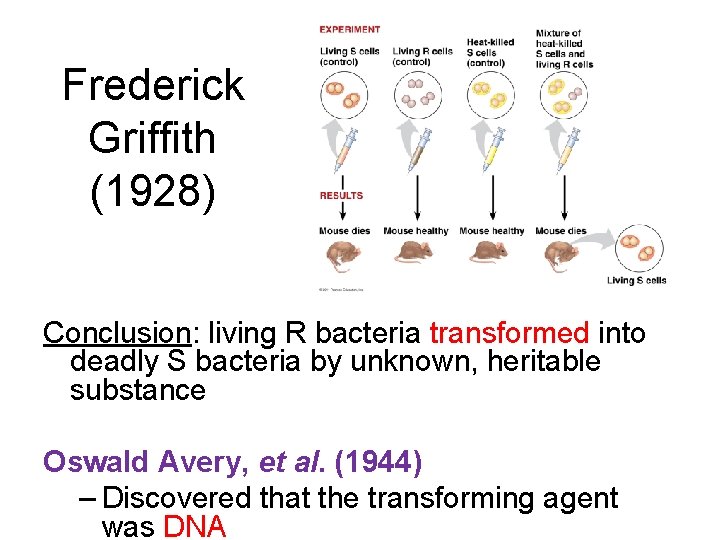
Frederick Griffith (1928) Conclusion: living R bacteria transformed into deadly S bacteria by unknown, heritable substance Oswald Avery, et al. (1944) – Discovered that the transforming agent was DNA
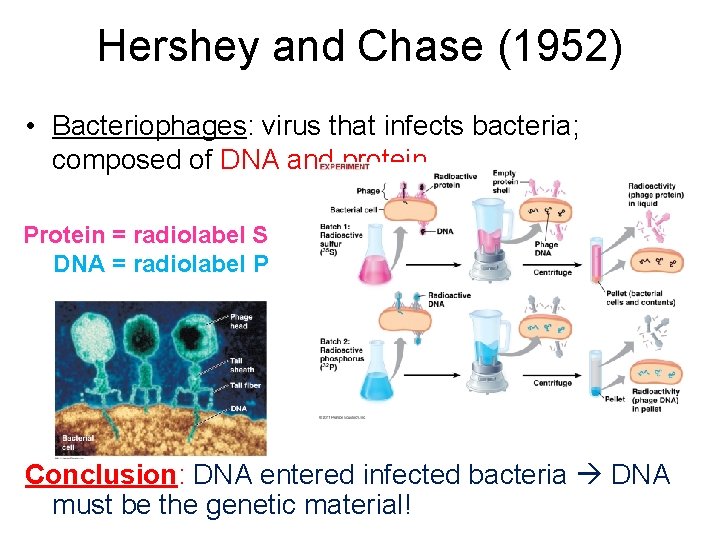
Hershey and Chase (1952) • Bacteriophages: virus that infects bacteria; composed of DNA and protein Protein = radiolabel S DNA = radiolabel P Conclusion: DNA entered infected bacteria DNA must be the genetic material!
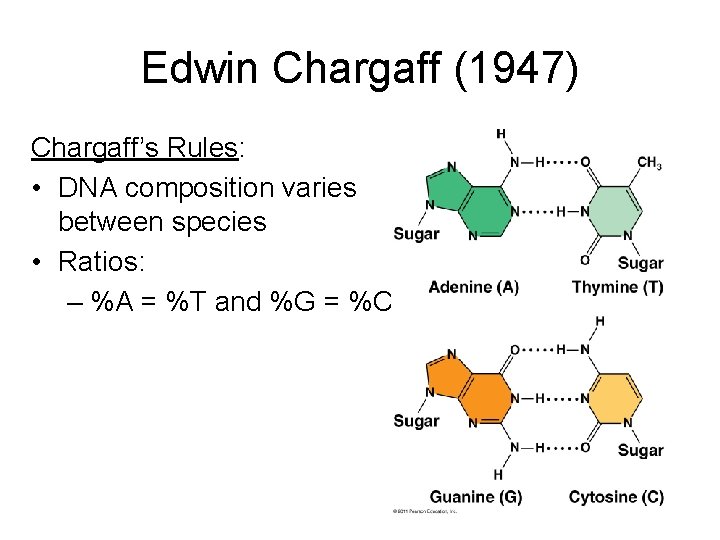
Edwin Chargaff (1947) Chargaff’s Rules: • DNA composition varies between species • Ratios: – %A = %T and %G = %C
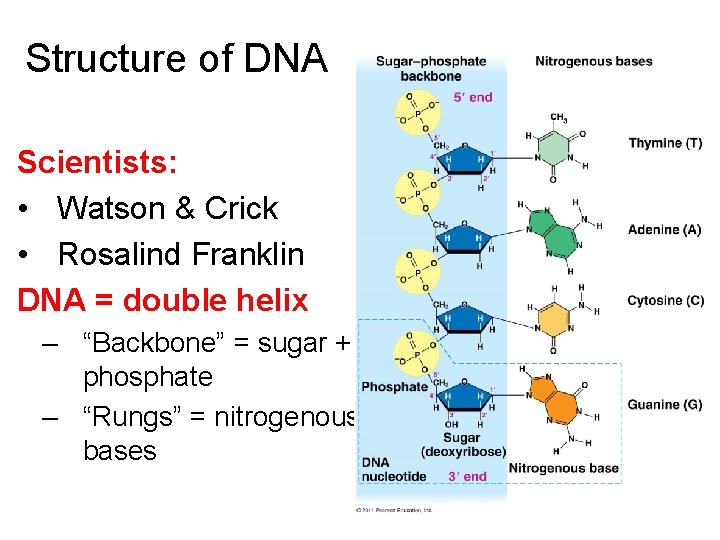
Structure of DNA Scientists: • Watson & Crick • Rosalind Franklin DNA = double helix – “Backbone” = sugar + phosphate – “Rungs” = nitrogenous bases
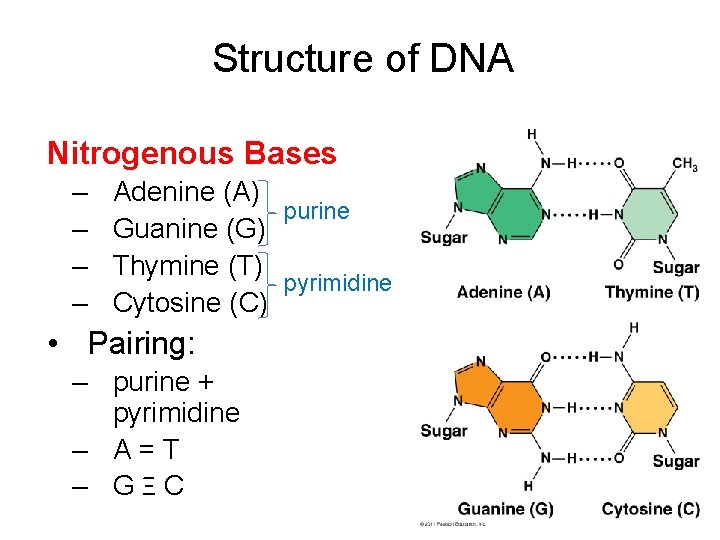
Structure of DNA Nitrogenous Bases – – Adenine (A) purine Guanine (G) Thymine (T) pyrimidine Cytosine (C) • Pairing: – purine + pyrimidine – A=T – GΞC
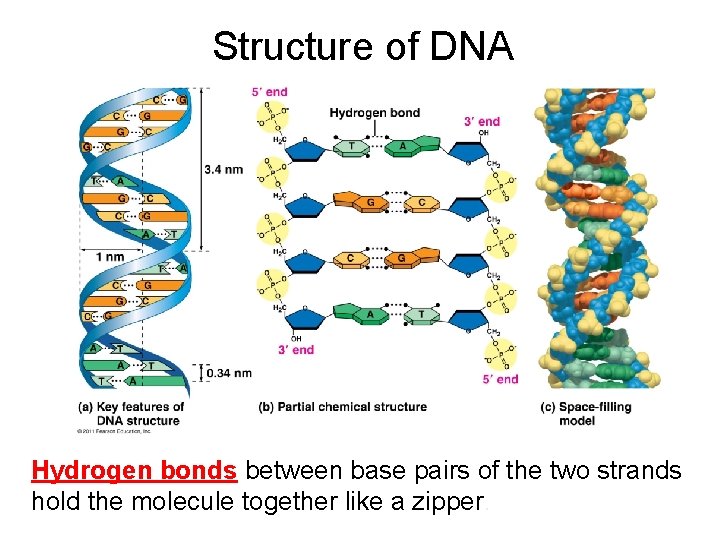
Structure of DNA Hydrogen bonds between base pairs of the two strands hold the molecule together like a zipper.
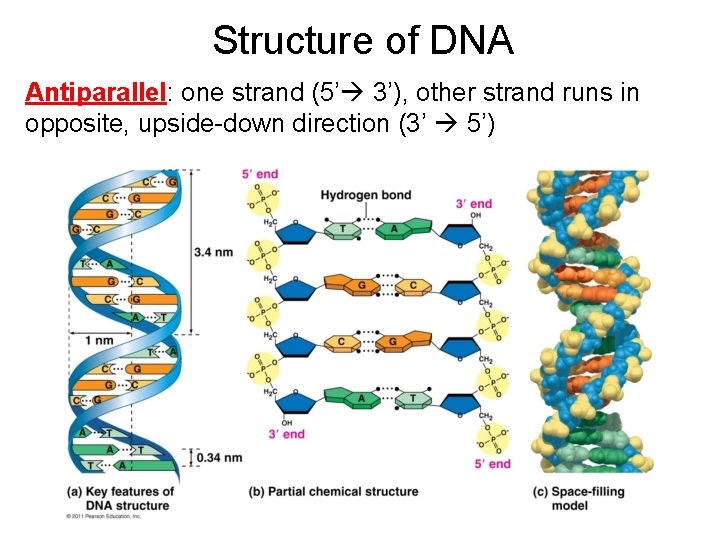
Structure of DNA Antiparallel: one strand (5’ 3’), other strand runs in opposite, upside-down direction (3’ 5’)
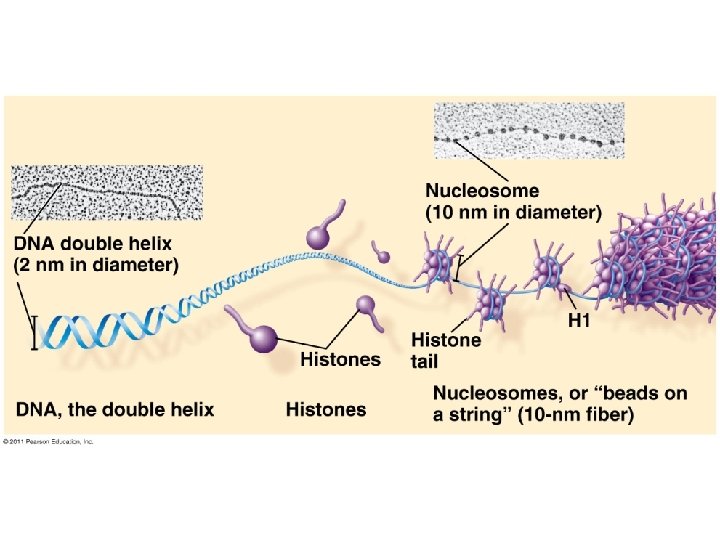

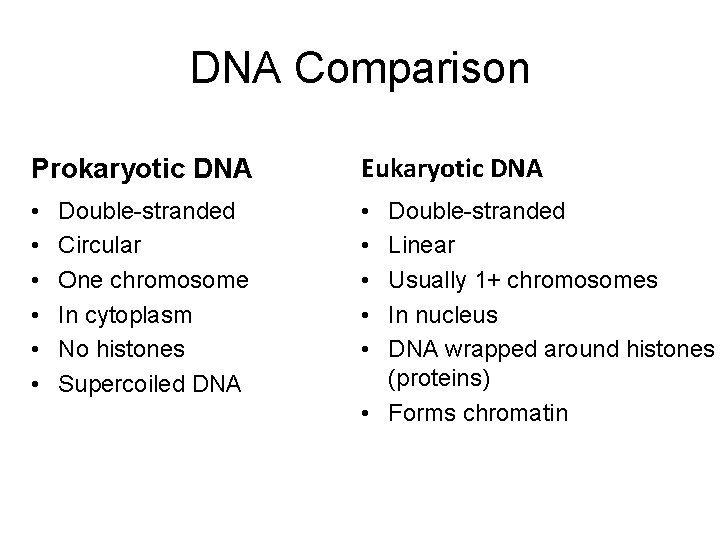
DNA Comparison Prokaryotic DNA Eukaryotic DNA • • • Double-stranded Circular One chromosome In cytoplasm No histones Supercoiled DNA Double-stranded Linear Usually 1+ chromosomes In nucleus DNA wrapped around histones (proteins) • Forms chromatin
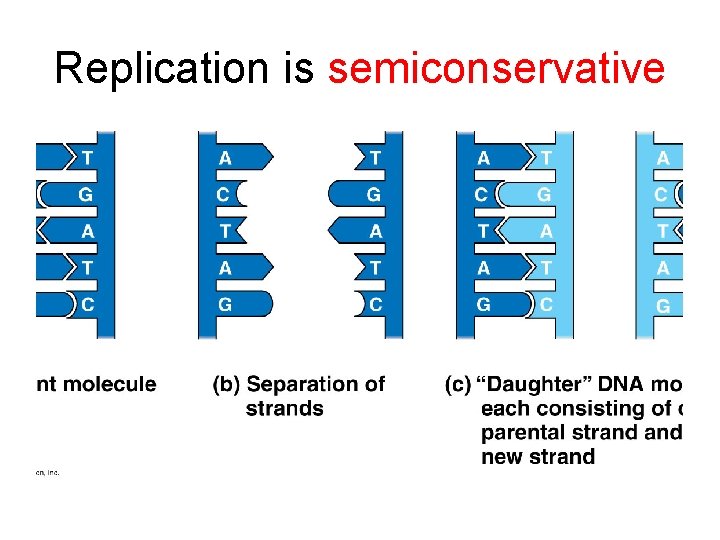
Replication is semiconservative
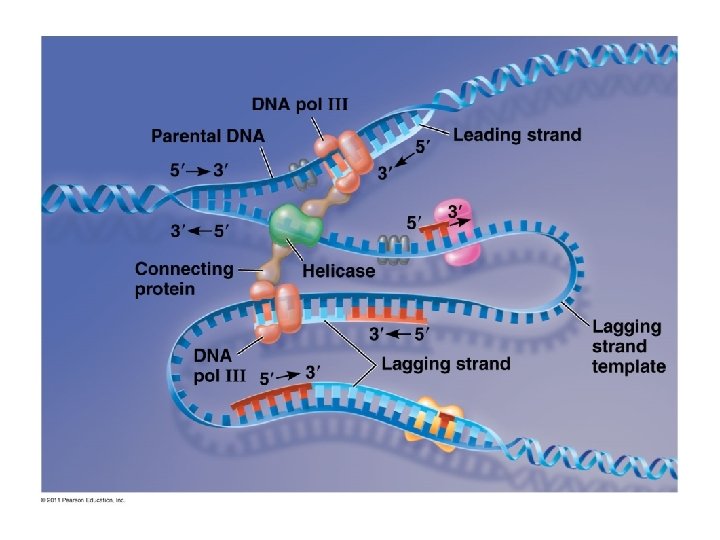

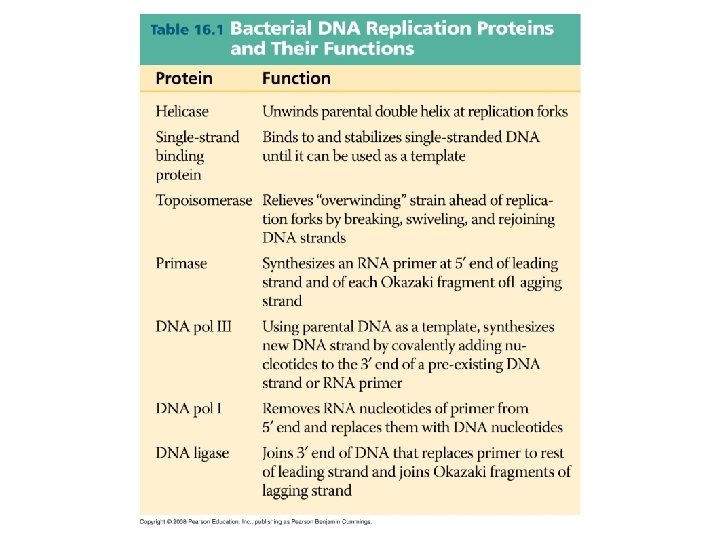
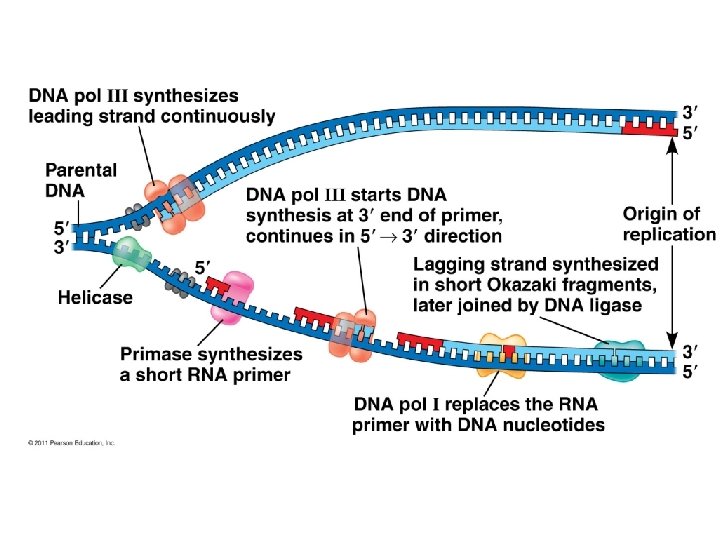
Major Steps of Replication: 1. Helicase: unwinds DNA at origins of replication 2. Initiation proteins separate 2 strands forms replication bubble 3. Primase: puts down RNA primer to start replication 4. DNA polymerase III: adds complimentary bases to leading strand (new DNA is made 5’ 3’) 5. Lagging strand grows in 3’ 5’ direction by the addition of Okazaki fragments 6. DNA polymerase I: replaces RNA primers with DNA 7. DNA ligase: seals fragments together
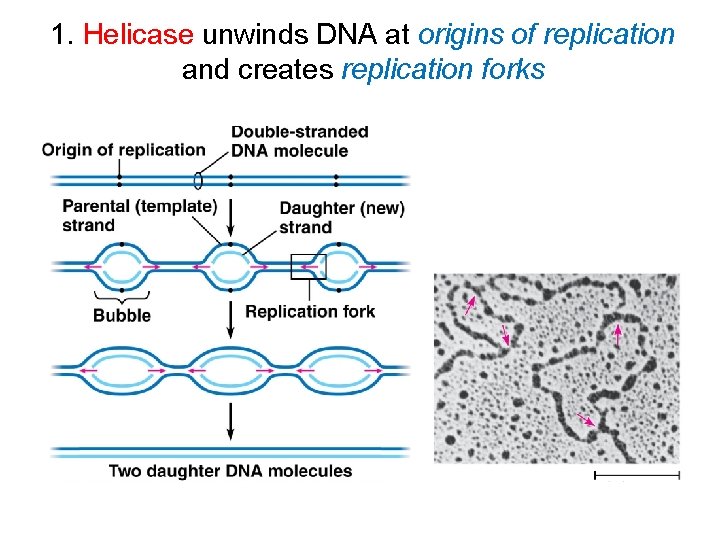
1. Helicase unwinds DNA at origins of replication and creates replication forks
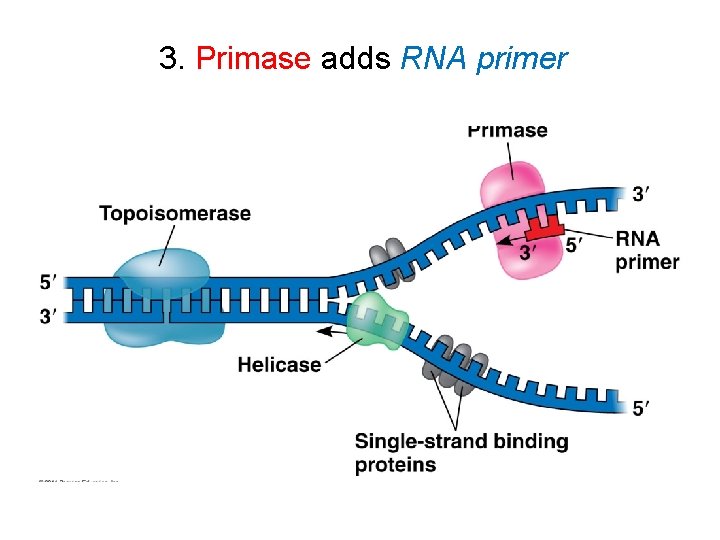
3. Primase adds RNA primer
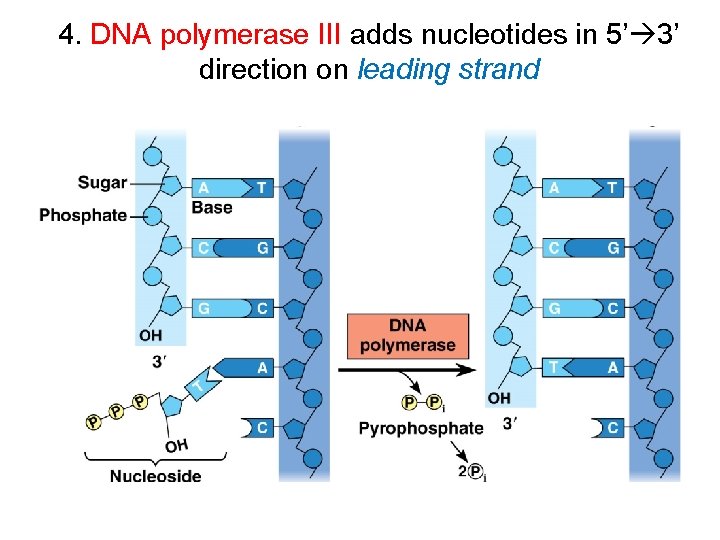
4. DNA polymerase III adds nucleotides in 5’ 3’ direction on leading strand
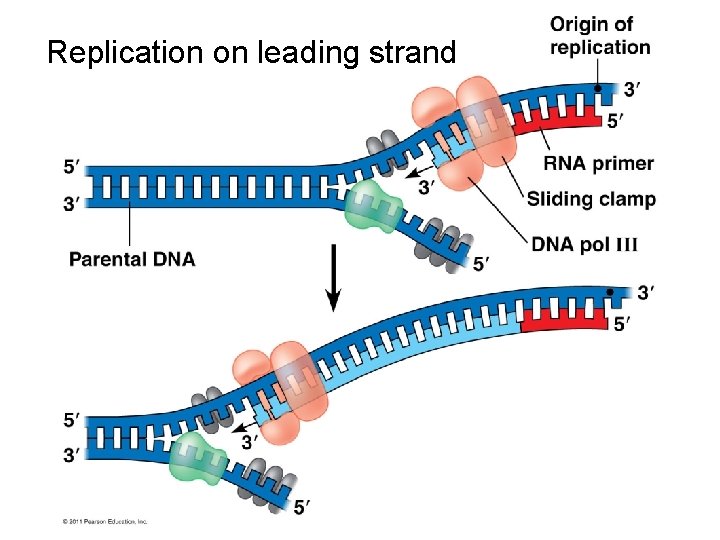
Replication on leading strand
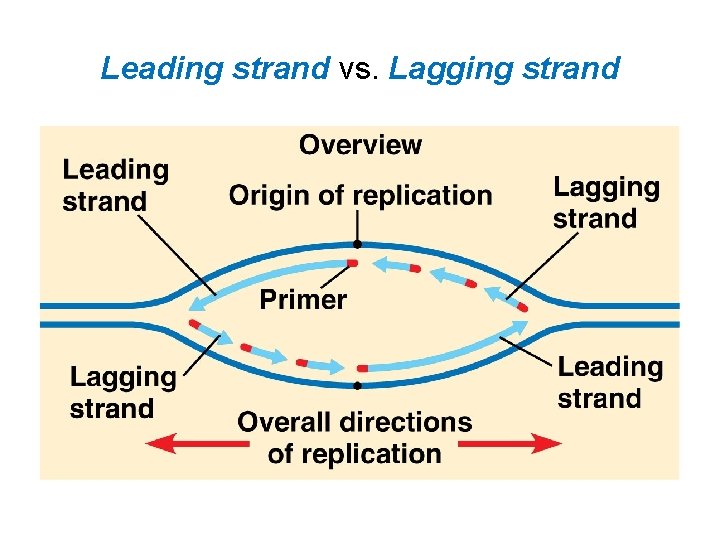
Leading strand vs. Lagging strand
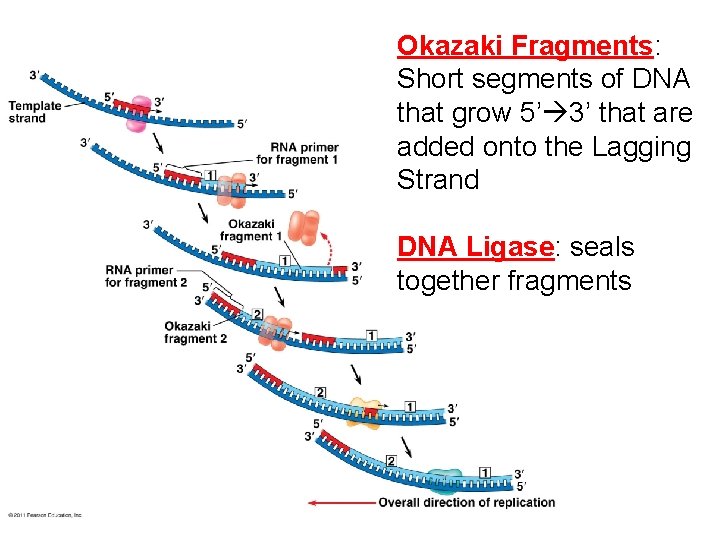
Okazaki Fragments: Short segments of DNA that grow 5’ 3’ that are added onto the Lagging Strand DNA Ligase: seals together fragments
Proofreading and Repair • DNA polymerases proofread as bases added • Mismatch repair: special enzymes fix incorrect pairings • Nucleotide excision repair: – Nucleases cut damaged DNA – DNA poly and ligase fill in gaps
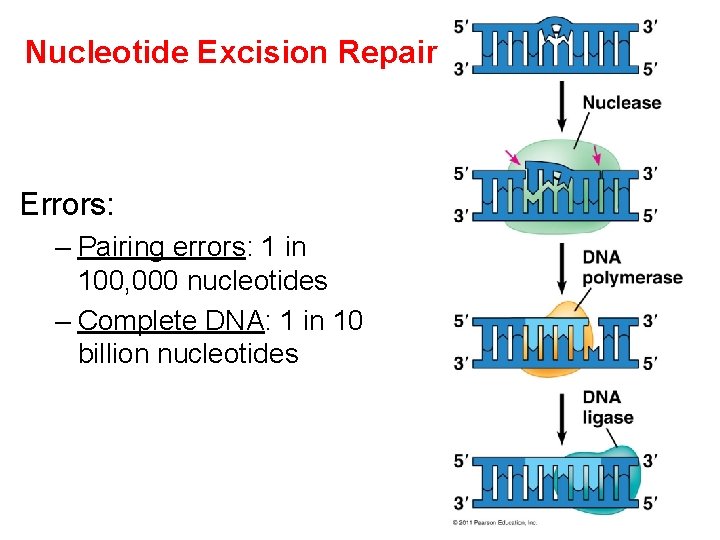
Nucleotide Excision Repair Errors: – Pairing errors: 1 in 100, 000 nucleotides – Complete DNA: 1 in 10 billion nucleotides
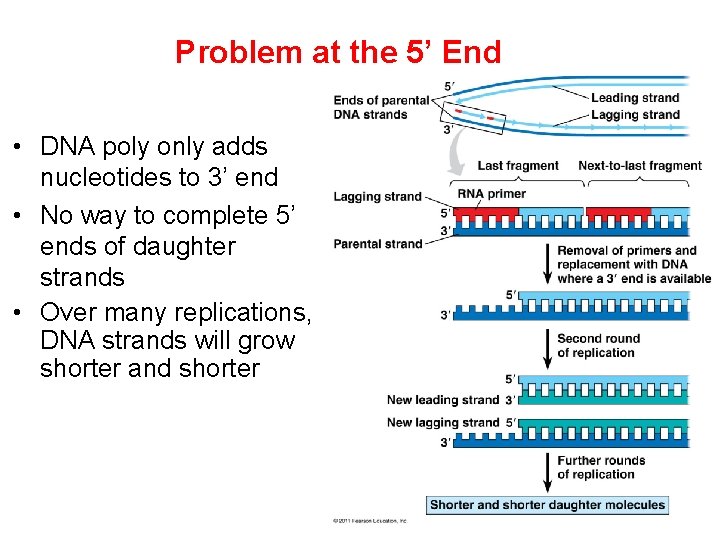
Problem at the 5’ End • DNA poly only adds nucleotides to 3’ end • No way to complete 5’ ends of daughter strands • Over many replications, DNA strands will grow shorter and shorter
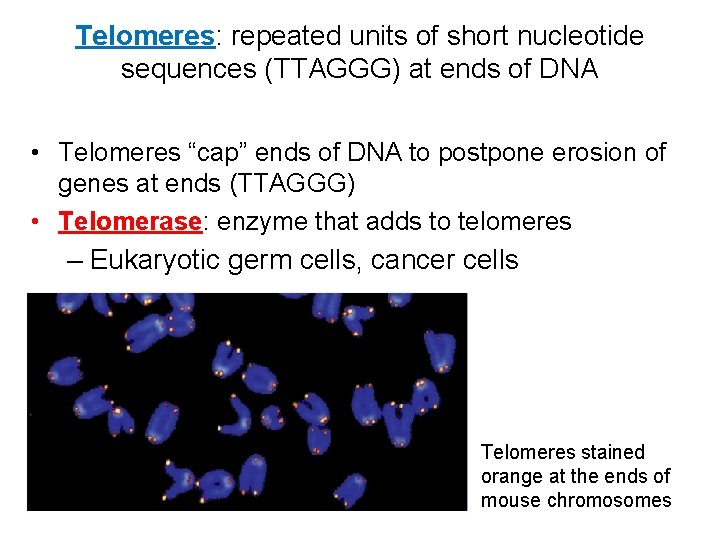
Telomeres: repeated units of short nucleotide sequences (TTAGGG) at ends of DNA • Telomeres “cap” ends of DNA to postpone erosion of genes at ends (TTAGGG) • Telomerase: enzyme that adds to telomeres – Eukaryotic germ cells, cancer cells Telomeres stained orange at the ends of mouse chromosomes
Flow of genetic information • Central Dogma: DNA RNA protein – Transcription: DNA RNA – Translation: RNA protein • Ribosome = site of translation • Gene Expression: process by which DNA directs the synthesis of proteins (or RNAs)
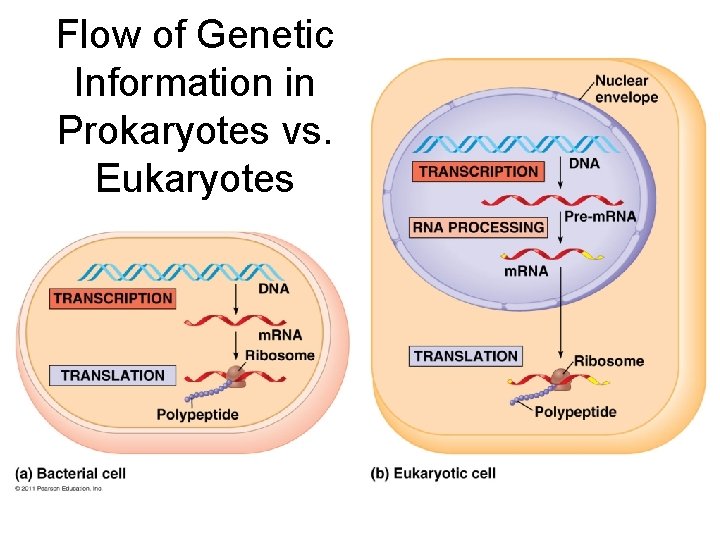
Flow of Genetic Information in Prokaryotes vs. Eukaryotes
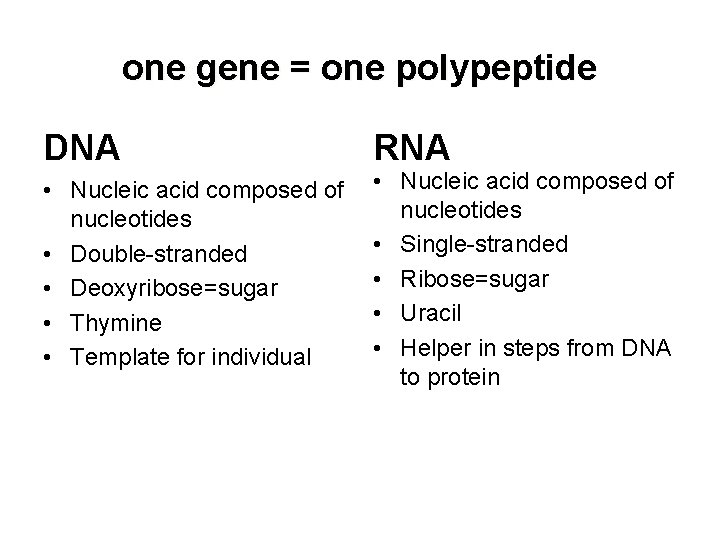
one gene = one polypeptide DNA • Nucleic acid composed of nucleotides • Double-stranded • Deoxyribose=sugar • Thymine • Template for individual RNA • Nucleic acid composed of nucleotides • Single-stranded • Ribose=sugar • Uracil • Helper in steps from DNA to protein
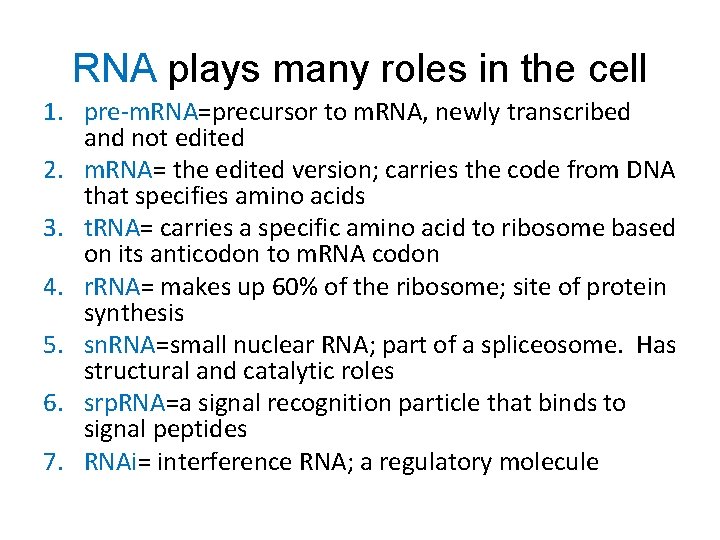
RNA plays many roles in the cell 1. pre-m. RNA=precursor to m. RNA, newly transcribed and not edited 2. m. RNA= the edited version; carries the code from DNA that specifies amino acids 3. t. RNA= carries a specific amino acid to ribosome based on its anticodon to m. RNA codon 4. r. RNA= makes up 60% of the ribosome; site of protein synthesis 5. sn. RNA=small nuclear RNA; part of a spliceosome. Has structural and catalytic roles 6. srp. RNA=a signal recognition particle that binds to signal peptides 7. RNAi= interference RNA; a regulatory molecule
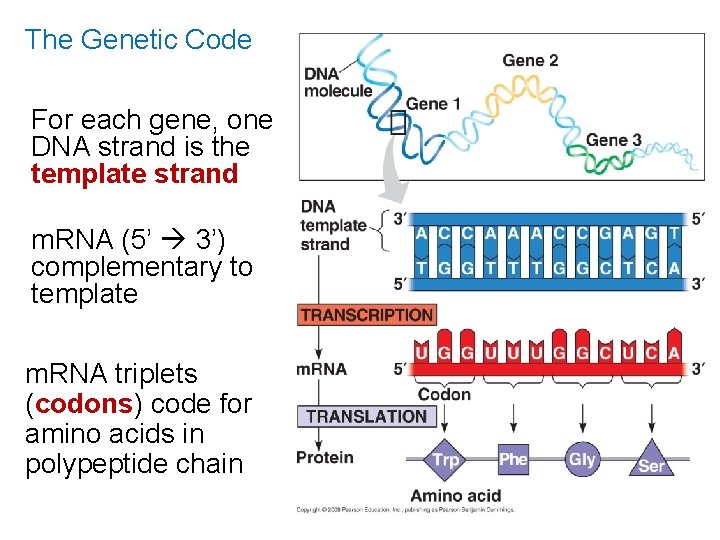
The Genetic Code For each gene, one DNA strand is the template strand m. RNA (5’ 3’) complementary to template m. RNA triplets (codons) code for amino acids in polypeptide chain
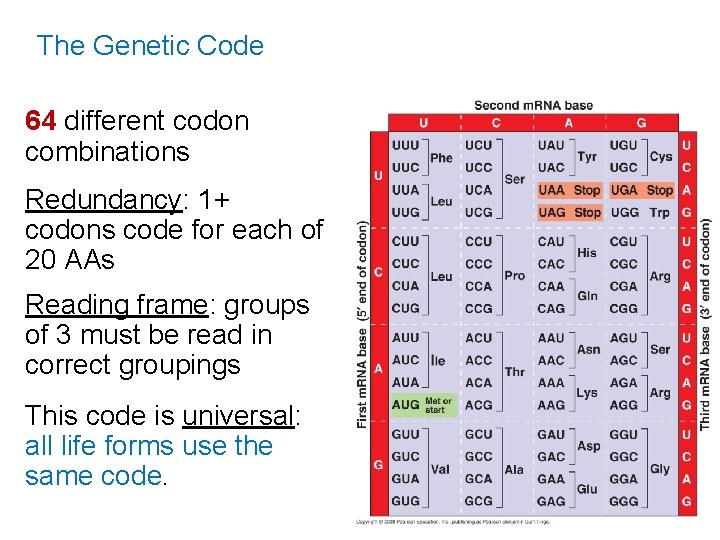
The Genetic Code 64 different codon combinations Redundancy: 1+ codons code for each of 20 AAs Reading frame: groups of 3 must be read in correct groupings This code is universal: all life forms use the same code.
Transcription unit: stretch of DNA that codes for a polypeptide or RNA (eg. t. RNA, r. RNA) RNA polymerase: polymerase – Separates DNA strands and transcribes m. RNA – m. RNA elongates in 5’ 3’ direction – Uracil (U) replaces thymine (T) when pairing to adenine (A) – Attaches to promoter (start of gene) and stops at terminator (end of gene)
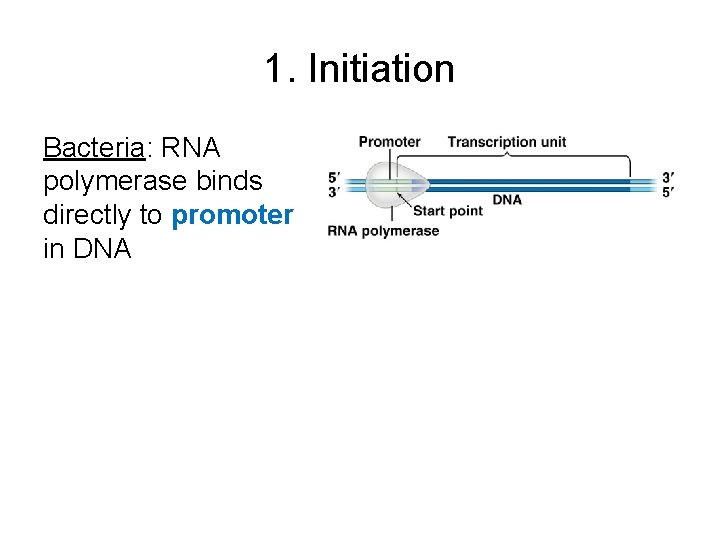
1. Initiation Bacteria: RNA polymerase binds directly to promoter in DNA
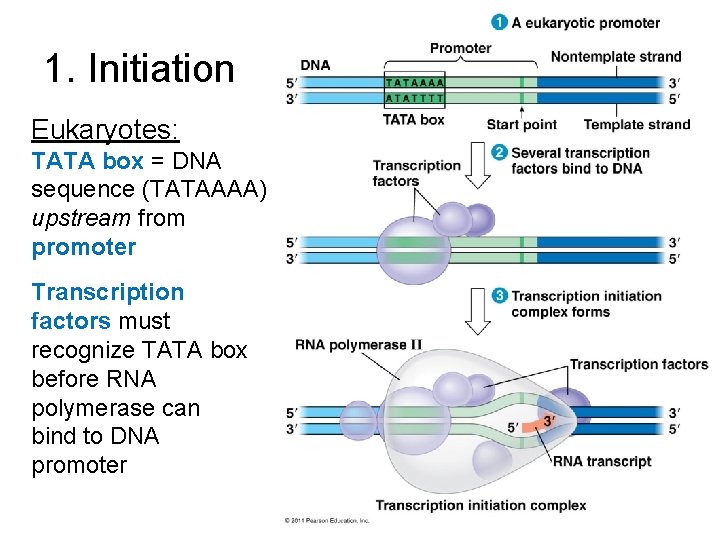
1. Initiation Eukaryotes: TATA box = DNA sequence (TATAAAA) upstream from promoter Transcription factors must recognize TATA box before RNA polymerase can bind to DNA promoter
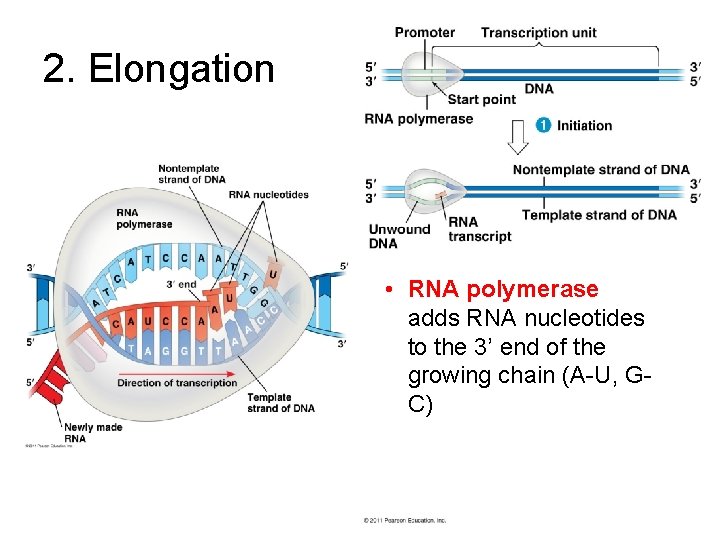
2. Elongation • RNA polymerase adds RNA nucleotides to the 3’ end of the growing chain (A-U, GC)
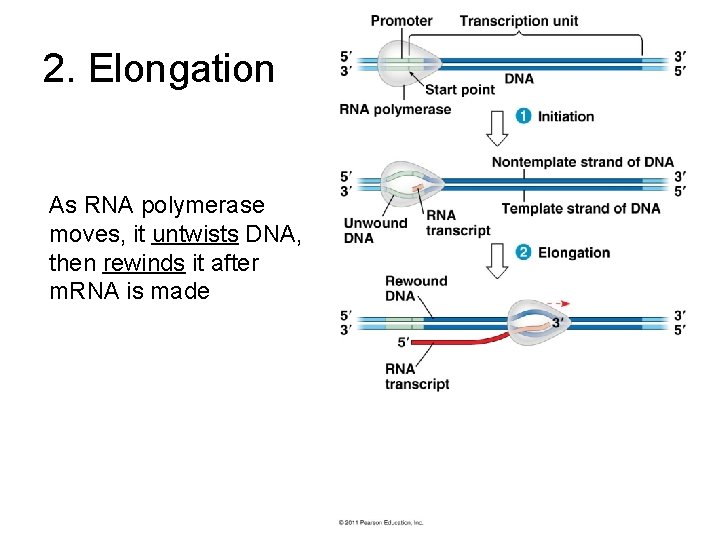
2. Elongation As RNA polymerase moves, it untwists DNA, then rewinds it after m. RNA is made
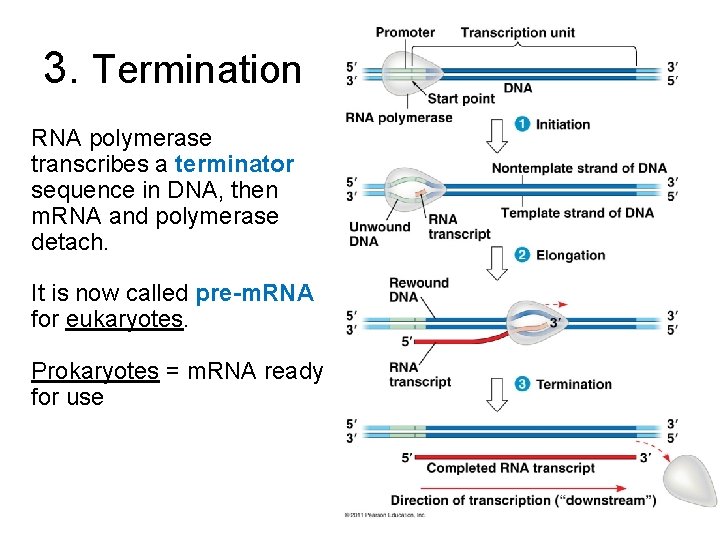
3. Termination RNA polymerase transcribes a terminator sequence in DNA, then m. RNA and polymerase detach. It is now called pre-m. RNA for eukaryotes. Prokaryotes = m. RNA ready for use
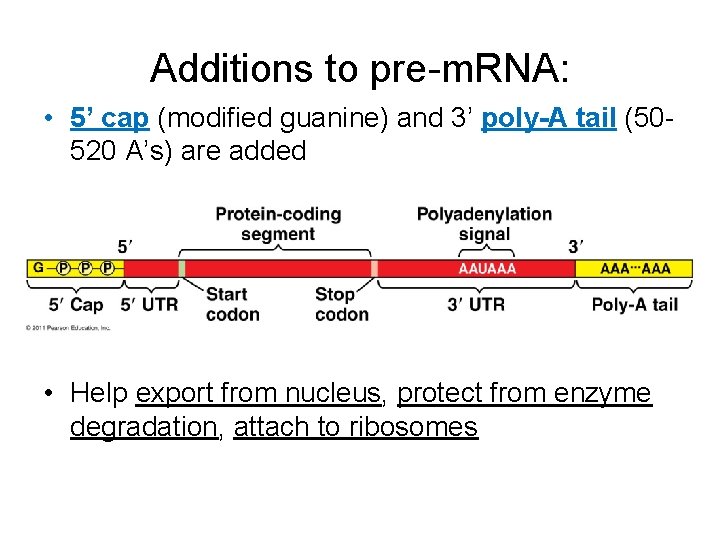
Additions to pre-m. RNA: • 5’ cap (modified guanine) and 3’ poly-A tail (50520 A’s) are added • Help export from nucleus, protect from enzyme degradation, attach to ribosomes
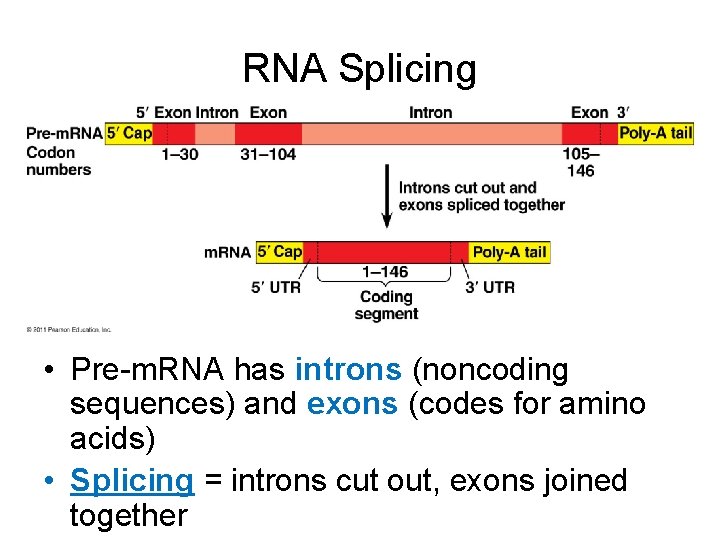
RNA Splicing • Pre-m. RNA has introns (noncoding sequences) and exons (codes for amino acids) • Splicing = introns cut out, exons joined together
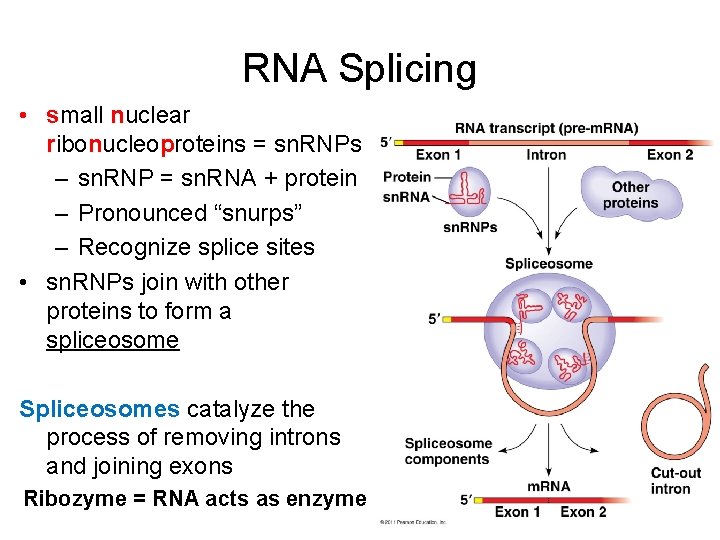
RNA Splicing • small nuclear ribonucleoproteins = sn. RNPs – sn. RNP = sn. RNA + protein – Pronounced “snurps” – Recognize splice sites • sn. RNPs join with other proteins to form a spliceosome Spliceosomes catalyze the process of removing introns and joining exons Ribozyme = RNA acts as enzyme
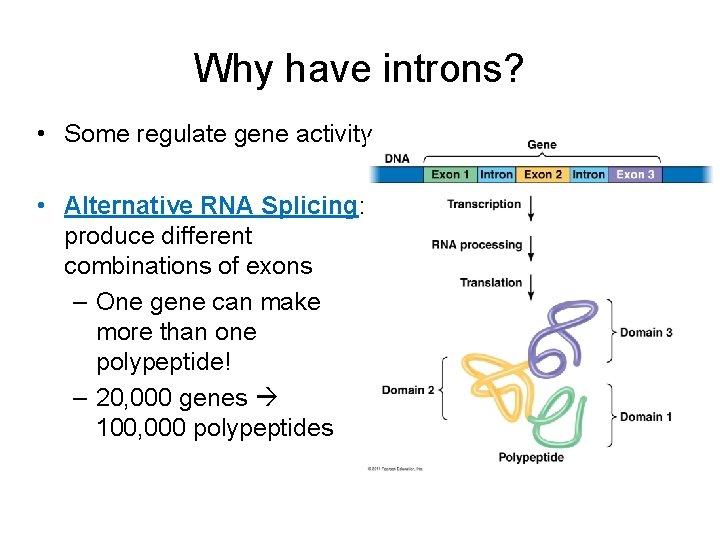
Why have introns? • Some regulate gene activity • Alternative RNA Splicing: produce different combinations of exons – One gene can make more than one polypeptide! – 20, 000 genes 100, 000 polypeptides
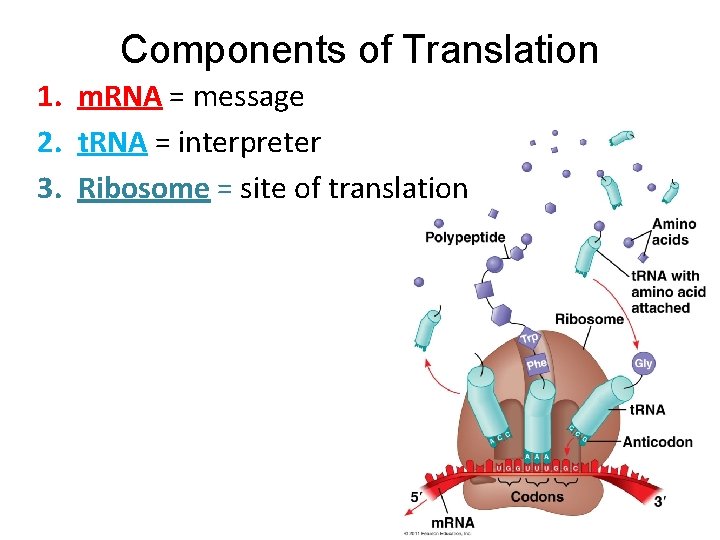
Components of Translation 1. m. RNA = message 2. t. RNA = interpreter 3. Ribosome = site of translation
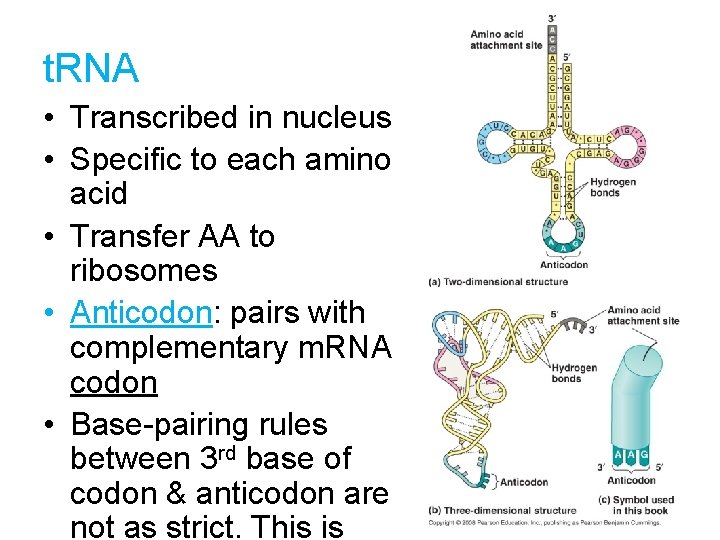
t. RNA • Transcribed in nucleus • Specific to each amino acid • Transfer AA to ribosomes • Anticodon: pairs with complementary m. RNA codon • Base-pairing rules between 3 rd base of codon & anticodon are not as strict. This is
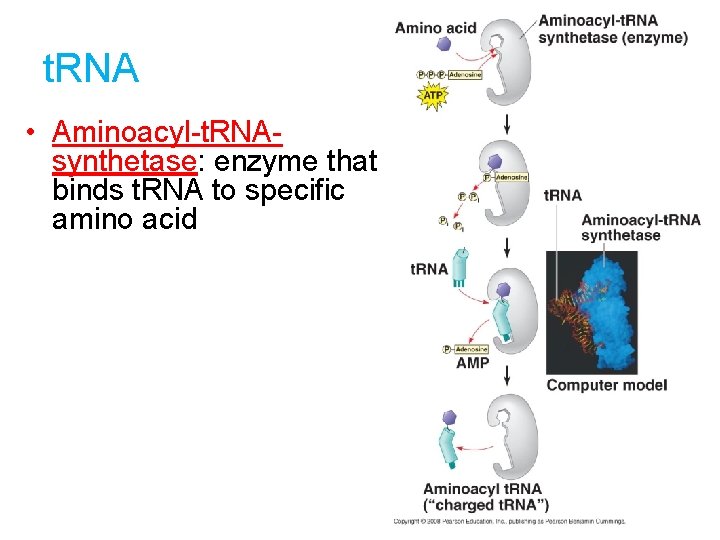
t. RNA • Aminoacyl-t. RNAsynthetase: enzyme that binds t. RNA to specific amino acid
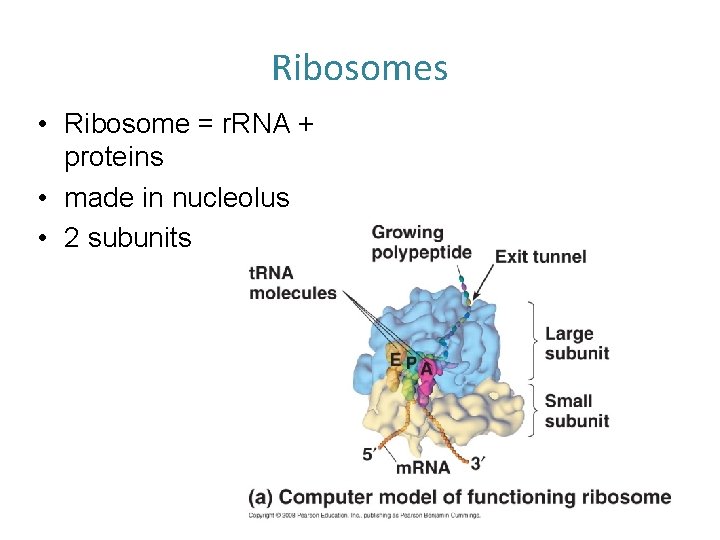
Ribosomes • Ribosome = r. RNA + proteins • made in nucleolus • 2 subunits
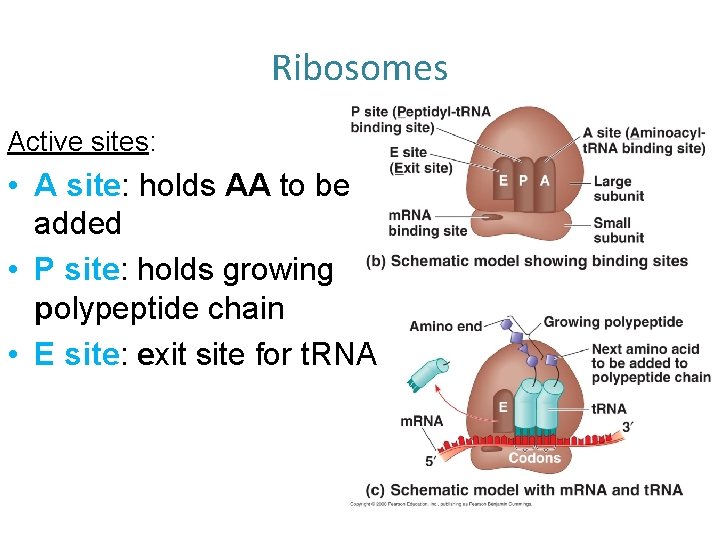
Ribosomes Active sites: • A site: holds AA to be added • P site: holds growing polypeptide chain • E site: exit site for t. RNA
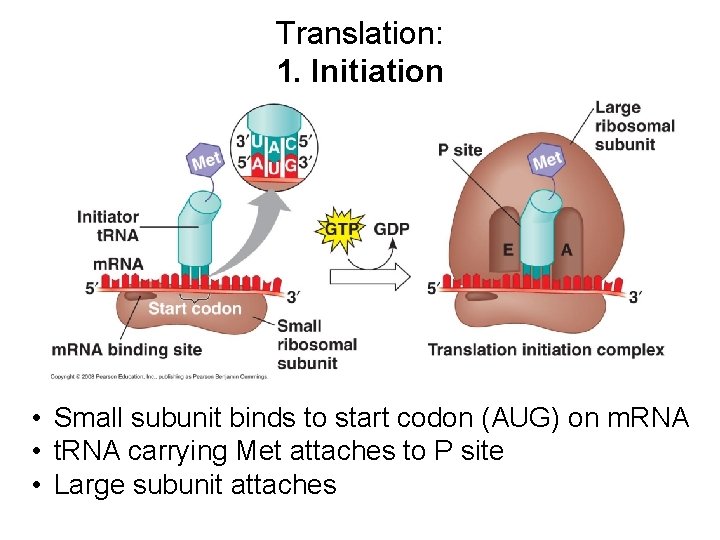
Translation: 1. Initiation • Small subunit binds to start codon (AUG) on m. RNA • t. RNA carrying Met attaches to P site • Large subunit attaches
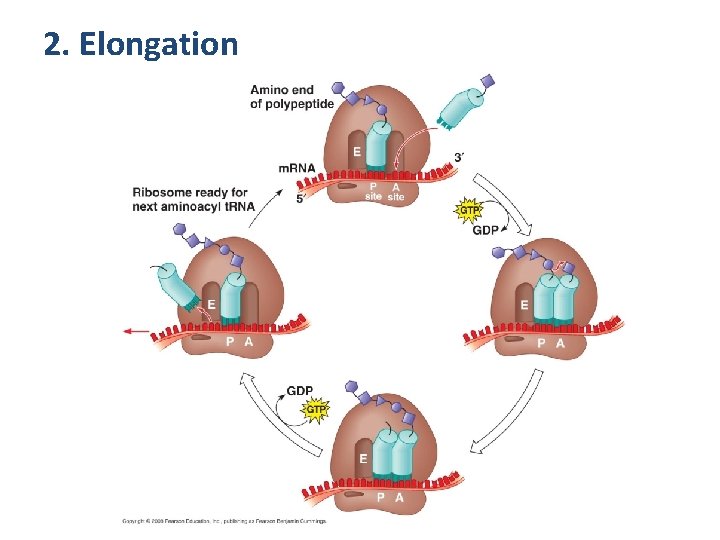
2. Elongation
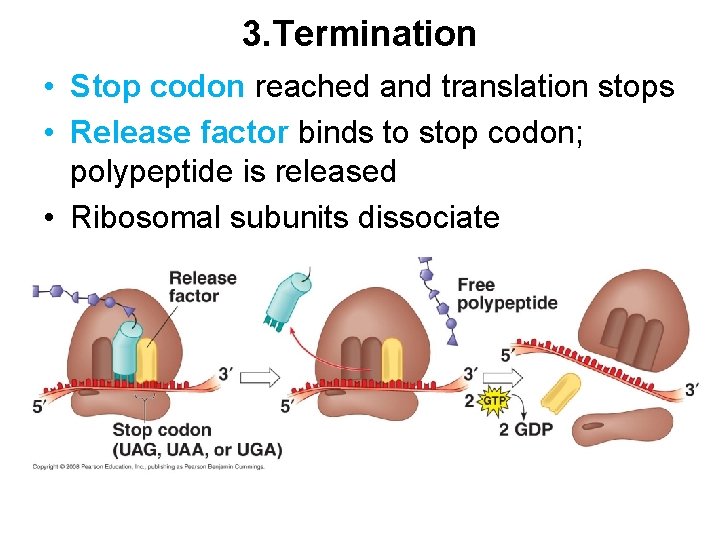
3. Termination • Stop codon reached and translation stops • Release factor binds to stop codon; polypeptide is released • Ribosomal subunits dissociate
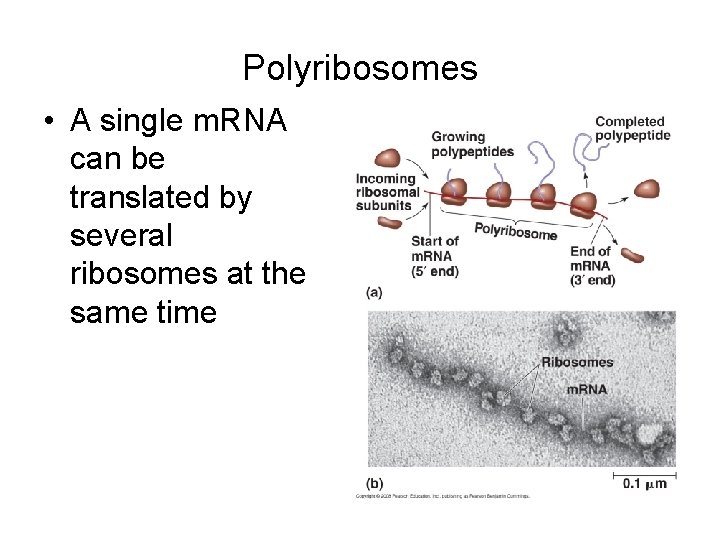
Polyribosomes • A single m. RNA can be translated by several ribosomes at the same time
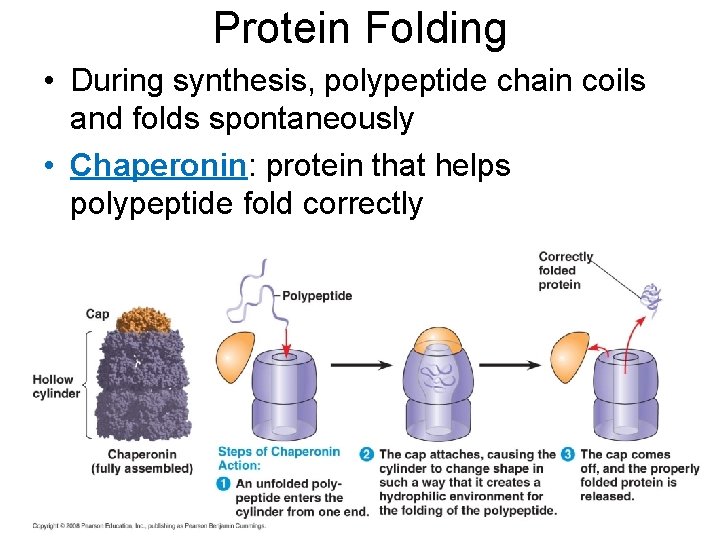
Protein Folding • During synthesis, polypeptide chain coils and folds spontaneously • Chaperonin: protein that helps polypeptide fold correctly
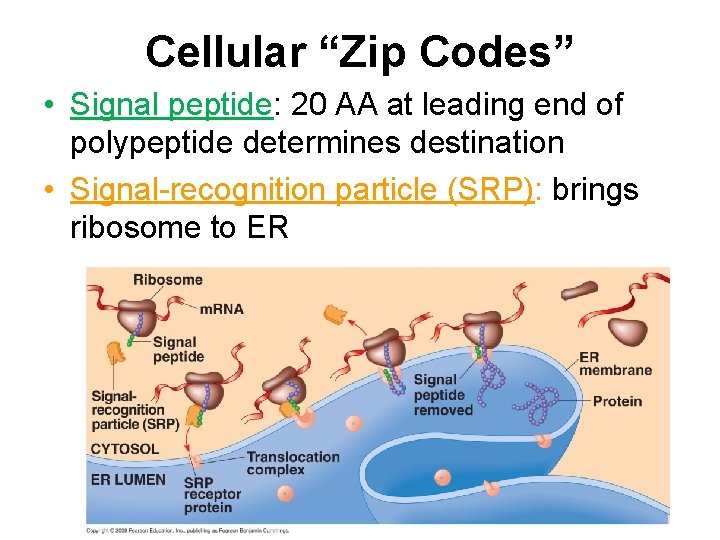
Cellular “Zip Codes” • Signal peptide: 20 AA at leading end of polypeptide determines destination • Signal-recognition particle (SRP): brings ribosome to ER
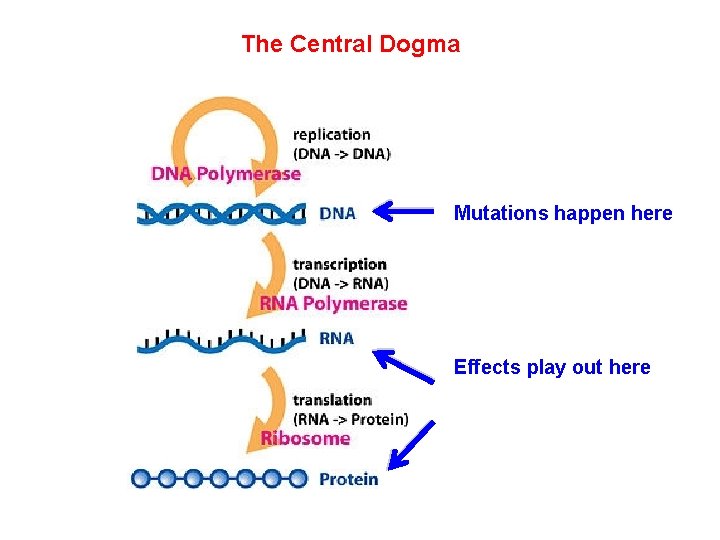
The Central Dogma Mutations happen here Effects play out here
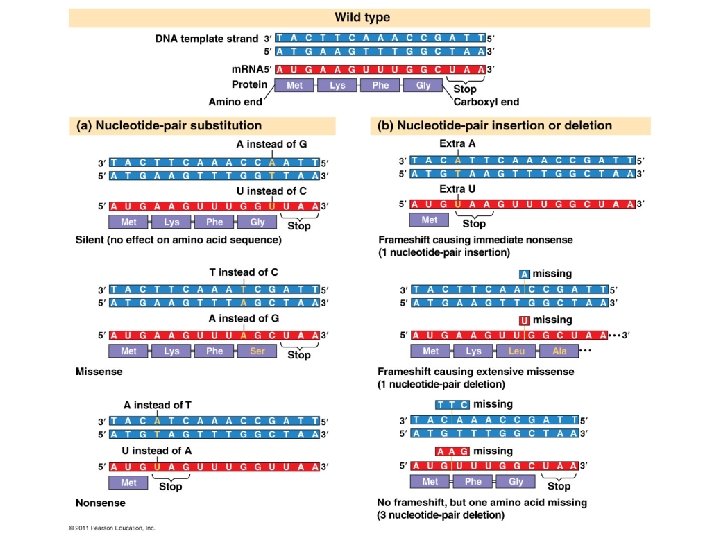
Mutations = changes in the genetic material of a cell • Large scale mutations: chromosomal; always cause disorders or death – nondisjunction, translocation, inversions, duplications, large deletions • Point mutations: alter 1 base pair of a gene 1. Base-pair substitutions – replace 1 with another • Missense: different amino acid • Nonsense: stop codon, not amino acid 2. Frameshift – m. RNA read incorrectly; nonfunctional proteins • Caused by insertions or deletions
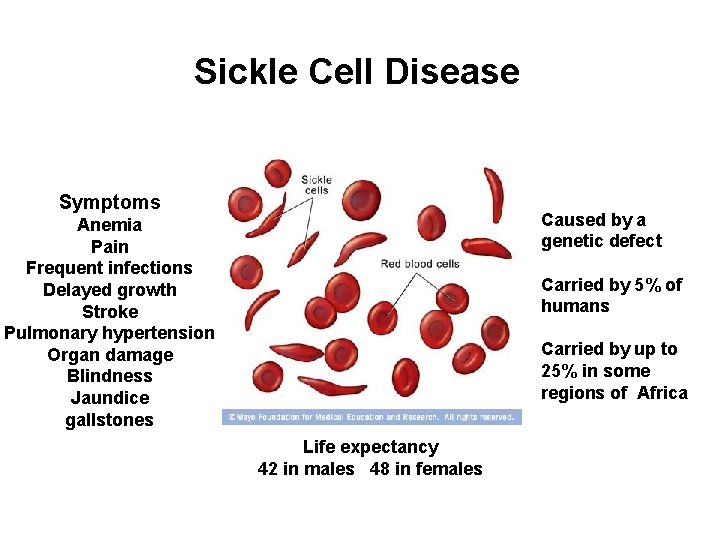
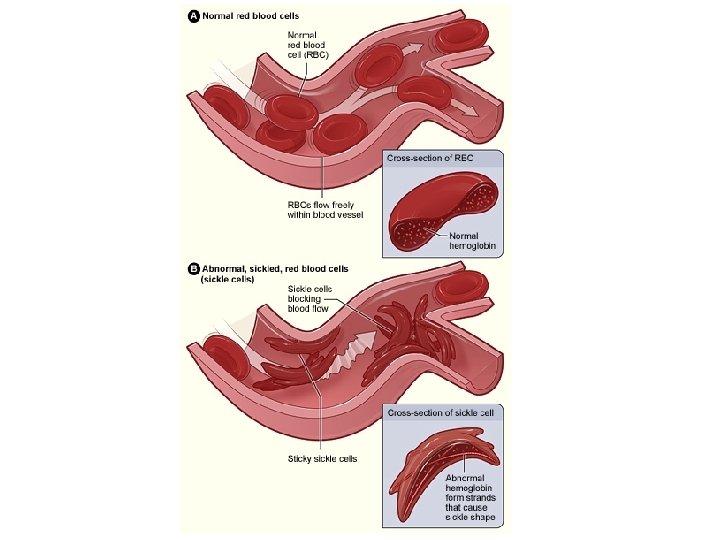
Sickle Cell Disease Symptoms Caused by a genetic defect Anemia Pain Frequent infections Delayed growth Stroke Pulmonary hypertension Organ damage Blindness Jaundice gallstones Carried by 5% of humans Carried by up to 25% in some regions of Africa Life expectancy 42 in males 48 in females
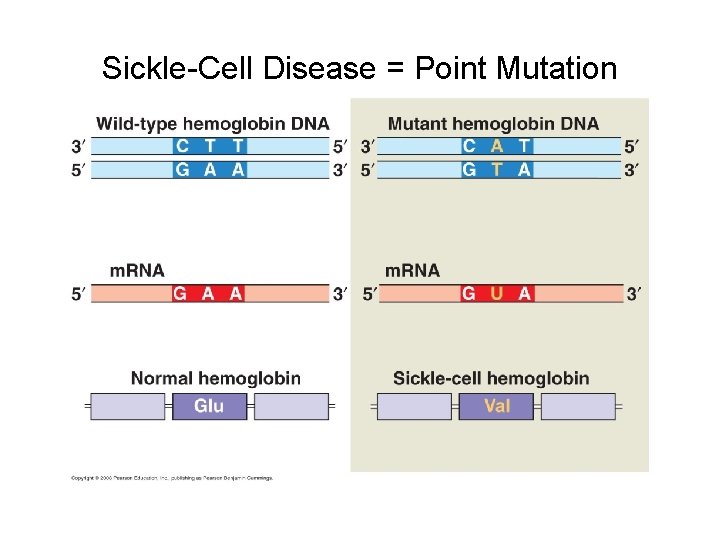
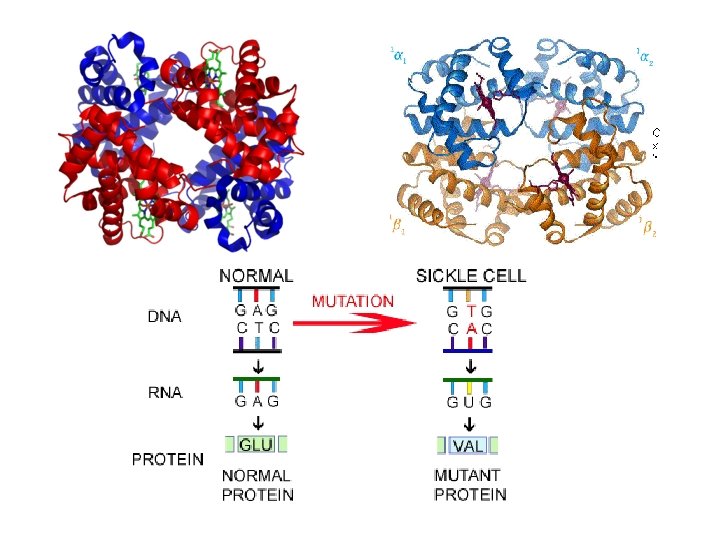
Sickle-Cell Disease = Point Mutation
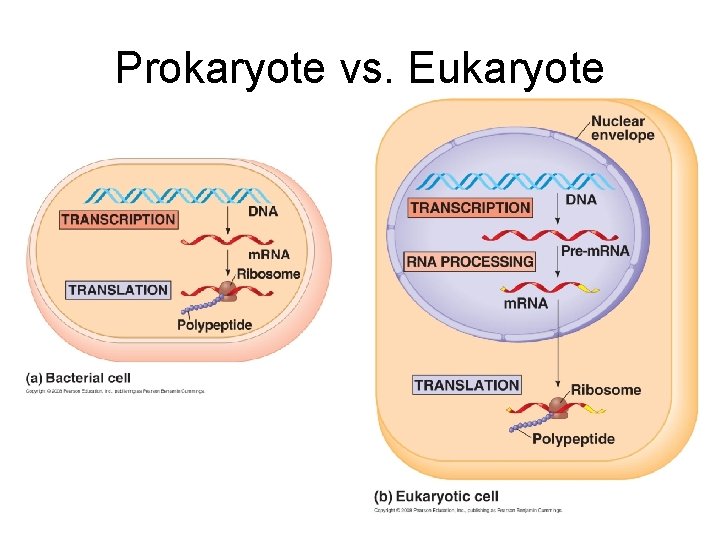
Prokaryote vs. Eukaryote
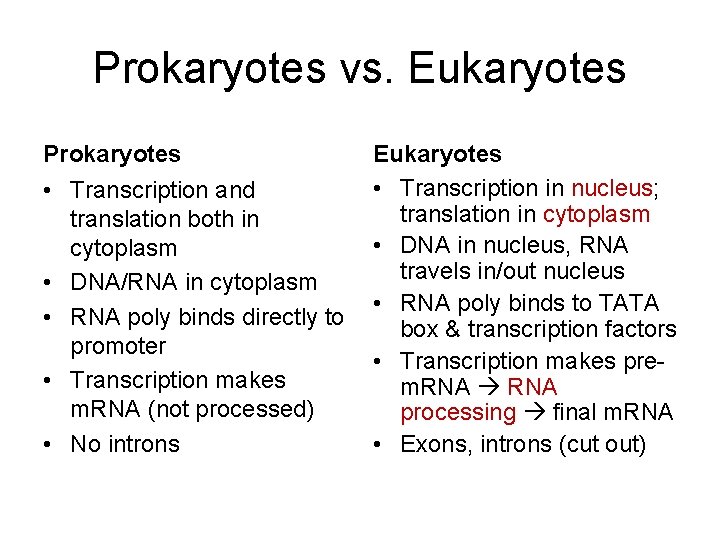
Prokaryotes vs. Eukaryotes Prokaryotes • Transcription and translation both in cytoplasm • DNA/RNA in cytoplasm • RNA poly binds directly to promoter • Transcription makes m. RNA (not processed) • No introns Eukaryotes • Transcription in nucleus; translation in cytoplasm • DNA in nucleus, RNA travels in/out nucleus • RNA poly binds to TATA box & transcription factors • Transcription makes prem. RNA processing final m. RNA • Exons, introns (cut out)
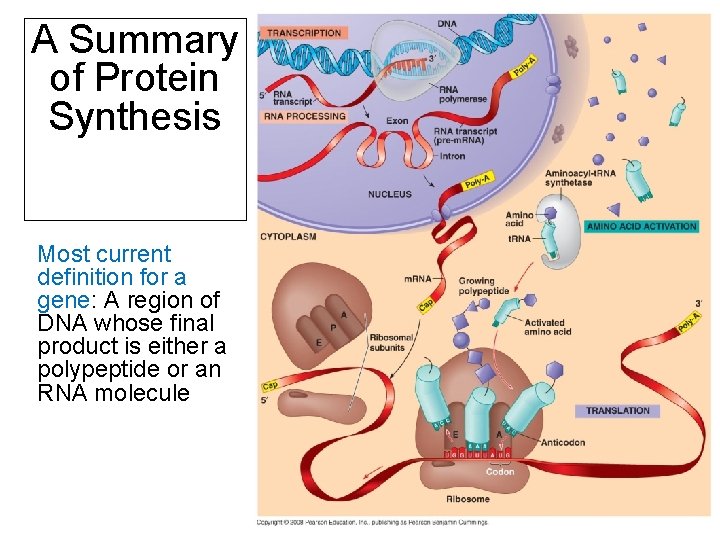
A Summary of Protein Synthesis Most current definition for a gene: A region of DNA whose final product is either a polypeptide or an RNA molecule
- Slides: 62