Forensics science Dr Nisreen tashkandy PEARSON 4 TH









- Slides: 28

Forensics science Dr Nisreen tashkandy PEARSON 4 TH EDITION 2019



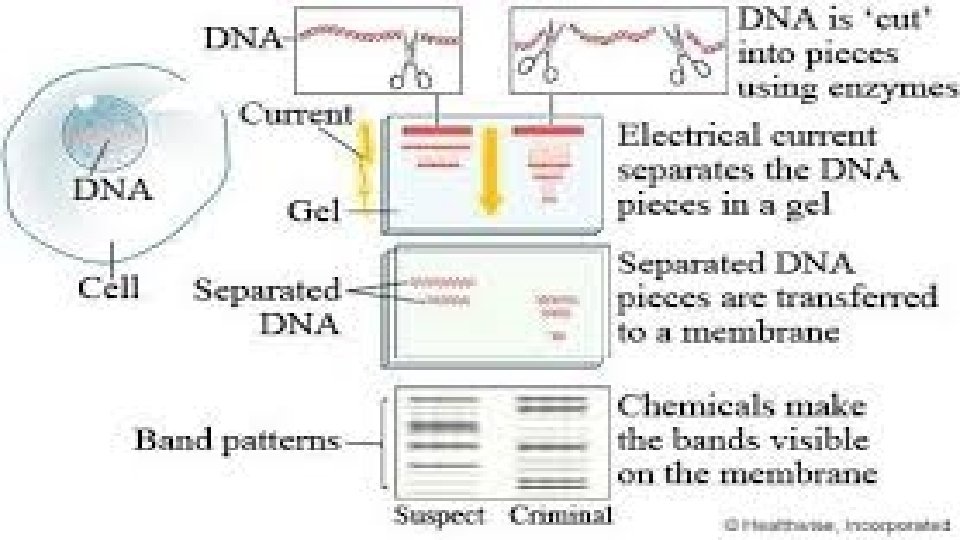
How Is DNA Typing Performed? • Only a tenth of 1 percent of DNA (about 3 million bases) differs from person to person. • These variable regions can generate a DNA profile of an individual, using samples from blood, bone, hair, or other body tissues containing a nucleus with DNA. • In criminal cases, this generally involves obtaining samples from crime scene evidence and a suspect, extracting the DNA, and analyzing it for the presence of a set of specific DNA regions.

How Is DNA Typing Performed? • There are two main types of forensic DNA testing: one based on restriction fragment length polymorphism (RFLP) • The polymerase chain reaction (PCR).

RFLP testing, requires • larger samples than does PCR, and the DNA must be intact and undegraded (the FBI accepts RFLPs of four types for case comparison). • view the animation “Restriction Fragment Length Polymorphisms” on the Companion Website for a visual analysis of the process. • Crime scene evidence that is old or present only in small amounts is often unsuitable for RFLP testing. Warm, moist conditions may accelerate DNA degradation, rendering it unsuitable for RFLP testing in a relatively short period of time. Because of this, RFLP is not used as often for DNA testing as it once was.

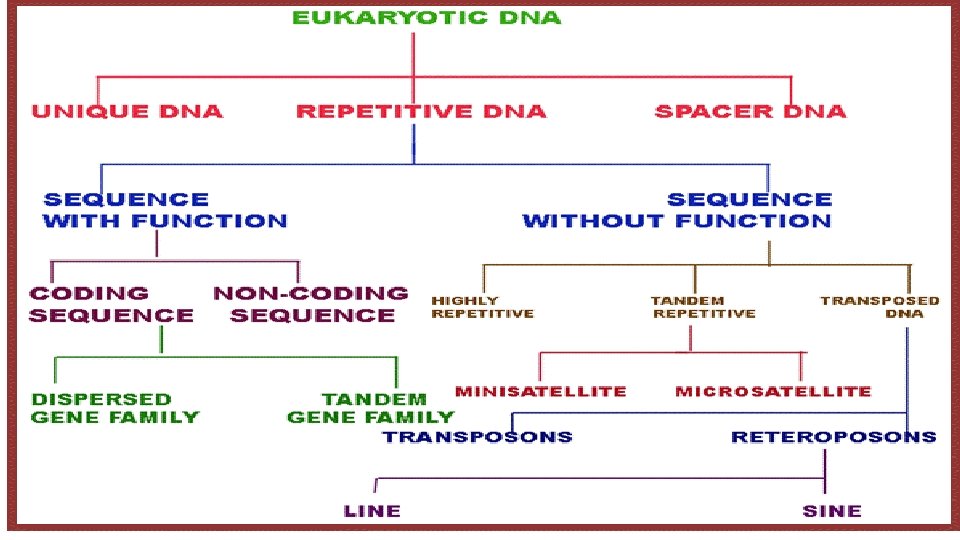
variable number tandem repeats (VNTRs) • Every strand of DNA is composed of both active genetic information, which codes proteins, and inactive DNA. • inactive portions contain repeated sequences of between 1 and 100 base pairs. • Every person has some VNTRs that were inherited from his or her mother and father. • No person has VNTRs that are identical to those of either parent (since we receive one allele from each parent, and they may not be the same). The individual’s VNTRs are a combination of repeats of those of theparents’ DNA regions in tandem.


variable number tandem repeats (VNTRs) • VNTRs are broken into two subgroups, the minisatellites, which are the basis for much of the RFLP analysis. • Microsatellites, which are the basis of the current CODIS data-base. • The uniqueness of an individual’s VNTRs provides the scientific marker of identity known as a DNA fingerprint.



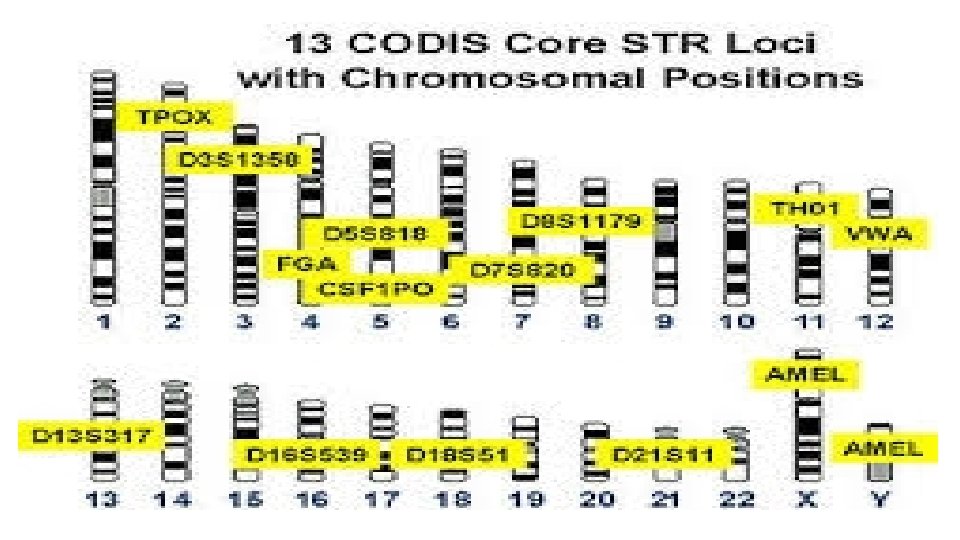
Short tandem repeat, or STR • DNA fingerprints produced by PCR are usually restricted to detecting the presence of microsatellites which are one-to six-nucleotide repeats dispersed throughout chromosomes. • Because these repeated regions can occur in many locations within the DNA, the probes used to identify them complement the DNA regions that surround the specific microsatellite being analyzed. • The FBI has chosen 13 unique STRs to test for DNA profiles contained within their Combined DNA Index System, or CODIS. • In early 2015, the FBI announced that an additional seven loci would be added to the CODIS Core effective January 1, 2017.

Preparing a DNA Fingerprint Preparing • Specimen collection. • DNA isolation and quantification. • and PCR amplification.

Specimen Collection Crime scene • Sunlight and high temperatures can degrade DNA. • Bacteria, busy doing their natural work as decomposers, can contaminate a sample before or during collection. • A small amount of moisture can damage and ruin a sample. DNA fingerprinting is a comparative process. • DNA from the crime scene must be compared with known DNA samples from the suspect. • DNA has been retrieved and successfully analyzed from samples that were a decade old by amplifying the DNA used for comparison by PCR.

Extracting DNA • for determining detergents that chemically wash away the unwanted cellular material. • followed by protein-and RNA-digesting enzymes and precipitation in ethanol. • Next, the DNA must be quantified for accurate comparisons with other samples. • Once the DNA is extracted and quantified, the technician must follow several steps to transform the unique signature of that DNA into visible evidence

PCR and STR Analysis • PCR is much like a photocopier for DNA. • locating the portions of DNA that can be useful for comparison. • Primers, or short pieces of DNA, are the tools used to find those portions on each DNA strand in the 5 to 3 direction. • The primers (about 20 nucleotides long) complement by DNA base-pairing interactions and bind best when the bases match exactly with complementary sections of the DNA.

PCR • copying begins, using a device called a thermal cycler. • enzyme Taq polymerase and DNA nucleotides, cycles of heat and cold cause the DNA to separate and then replicate repeatedly. • Within hours, the DNA segments can be increased by billions. • That rapid growth presents a possible problem: PCR is very sensitive to contamination, and a small error in a field or laboratory procedure can result in the duplication of a contaminating DNA sample.

STR Analysis • After STRs are amplified by PCR, the alleles are separated and detected using capillary electrophoresis, which separates the lengths of amplified DNA, allowing the number of repeats in each of the two alleles on homologous chromosomes to be determined. • The number of repeats within an STR is referred to as an allele. • The large number of variations for each STR provides the variability that is needed to distinguish different individuals using DNA fingerprints.

STR known as D 7 S 820 • found on chromosome 7, contains between 5 and 16 repeats of GATA. • An individual with D 7 S 820 alleles 10 and 15, for example, would have inherited a copy of D 7 S 820 with 10 GATA repeats from one parent and a copy of D 7 S 820 with 15 GATA repeats from the other parent. • There are 12 different alleles for this STR, creating 78 different possible genotypes, or pairs of alleles, for this one site.


Putting DNA to Use • evidence collected from a crime scene is compared with evidence collected from a known source. • During evaluation of the samples, analysts are looking for alignment of the bands. • If the samples are from the same person (or from identical twins), the bands or dots should line up exactly. • All tests are based on exclusion. In other words, testing continues only until a difference is found. If no difference is found after a statistically acceptable amount of testing, the probability of a match

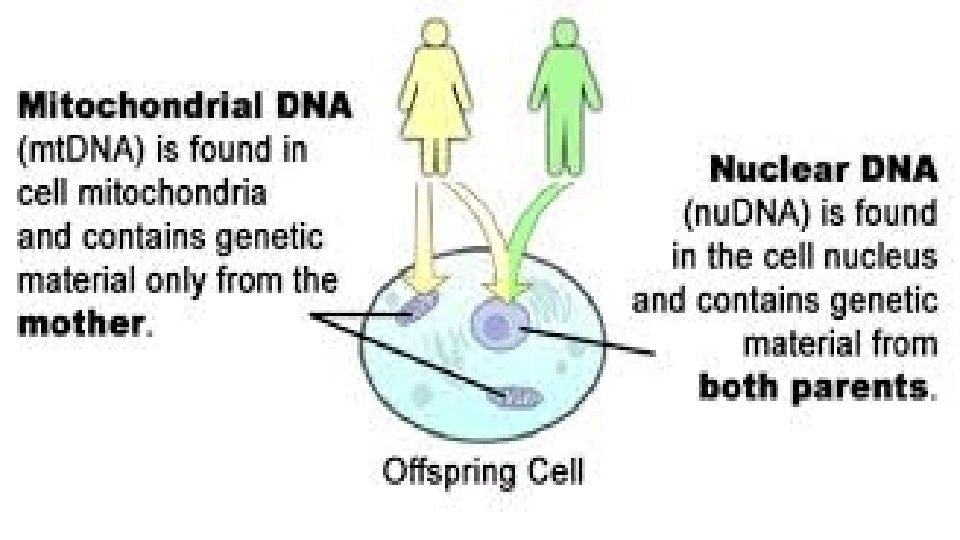
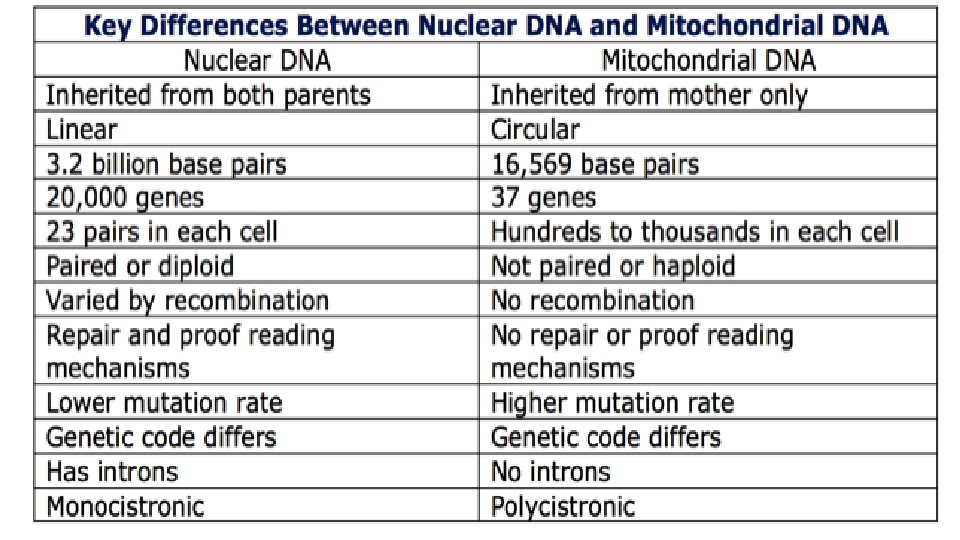
Mitochondrial DNA Analysis • analysis can be used to examine DNA from samples that cannot be analyzed by RFLP or STR. • DNA extracted from a mitochondrion. • The primary advantage of mt. DNA is that it is present in high copy number within cells and numerically more likely to be recovered from highly degraded specimens. • A major disadvantage is mt. DNA time-consuming to generate and review the 610 nucleotides of sequence information commonly targeted in hypervariable regions I and II (HVI and HVII) of the control region of the DNA. • All daughters have the same mt DNA as their mothers because the mitochondria of embryos come from the mother’s egg cell. The father’s sperm donates only nuclear DNA and no mitochondria.



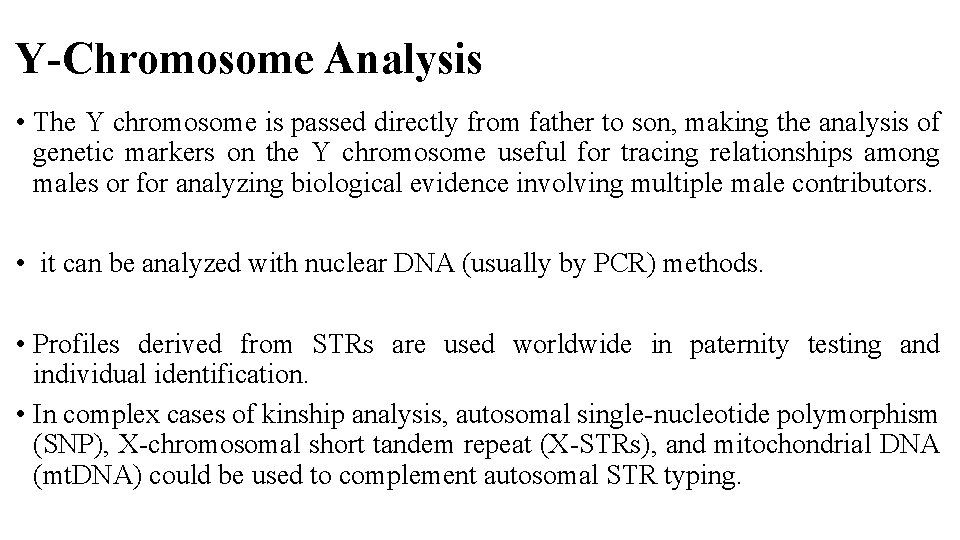
Y-Chromosome Analysis • The Y chromosome is passed directly from father to son, making the analysis of genetic markers on the Y chromosome useful for tracing relationships among males or for analyzing biological evidence involving multiple male contributors. • it can be analyzed with nuclear DNA (usually by PCR) methods. • Profiles derived from STRs are used worldwide in paternity testing and individual identification. • In complex cases of kinship analysis, autosomal single-nucleotide polymorphism (SNP), X-chromosomal short tandem repeat (X-STRs), and mitochondrial DNA (mt. DNA) could be used to complement autosomal STR typing.

Protein Analysis May Join DNA Forensics • Lawrence Livermore National Lab Forensic Science Center has used mass spectrometry–based shotgun proteomics to characterize hair shaft proteins in 66 European-American subjects. • People do not inherit proteins, but they do inherit the DNA that produces their protein. • Protein is not only chemically more robust than DNA, but it also contains genetic variation in the form of single amino acid polymorphisms (SAPs). • A total of 596 SNP alleles were correctly assigned in 32 loci from 22 genes of subjects’ DNA and directly validated using Sanger sequencing. • the protein identification method requires about 2 1/2 days.

One of the main goals of SNP research is to understand the genetics of the human phenotype variation • Human single nucleotide polymorphisms (SNPs) represent the most frequent type of human population DNA variation. • Non-synonymous coding SNPs (ns. SNPs) comprise a group of SNPs that, together with SNPs in regulatory regions, are believed to have the highest impact on phenotype. • database HGVbase, which gave rise to a dataset of ns. SNPs with predicted functionality.

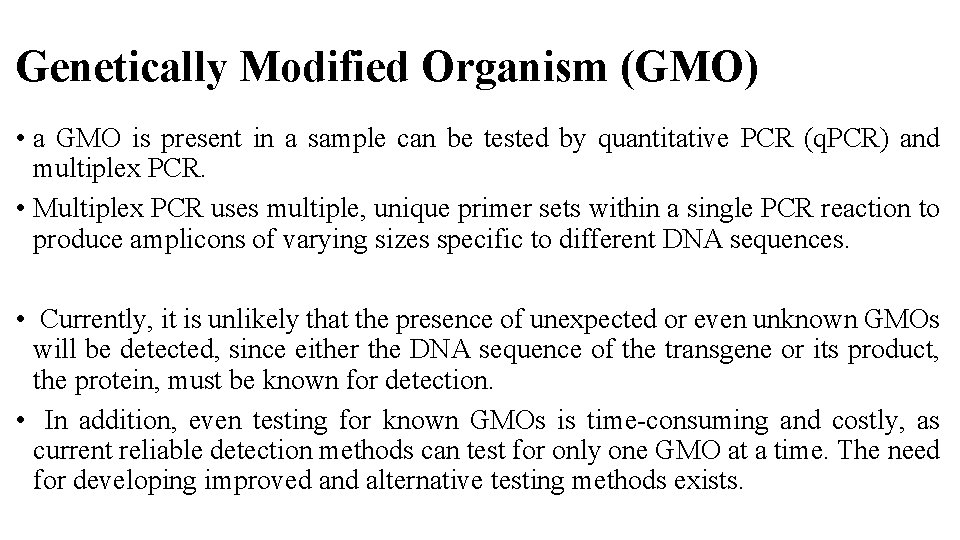
Genetically Modified Organism (GMO) • a GMO is present in a sample can be tested by quantitative PCR (q. PCR) and multiplex PCR. • Multiplex PCR uses multiple, unique primer sets within a single PCR reaction to produce amplicons of varying sizes specific to different DNA sequences. • Currently, it is unlikely that the presence of unexpected or even unknown GMOs will be detected, since either the DNA sequence of the transgene or its product, the protein, must be known for detection. • In addition, even testing for known GMOs is time-consuming and costly, as current reliable detection methods can test for only one GMO at a time. The need for developing improved and alternative testing methods exists.