First Aid Pathology Data quality assessment in PHENIX











- Slides: 31

First Aid & Pathology Data quality assessment in PHENIX Peter Zwart Computational Crystallography Initiative Physical Biosciences Division

Introduction • PHENIX: – Software for bio-molecular crystallography • • • Molecular replacement (PHASER) Substructure solution (SOLVE, HYSS) Phasing (SOLVE; PHASER) Model building (RESOLVE) Refinement (phenix. refine) Ligand building (ELBOW and RESOLVE) • http: //www. phenix-online. org Computational Crystallography Initiative Physical Biosciences Division

Introduction • GUI shapshots Computational Crystallography Initiative Physical Biosciences Division

Introduction • Structure solution can be enhanced by the knowledge of the quality of the merged data – – – Presence of absence of anomalous signal Completeness Twinning Anisotropy Pseudo centering … • Adapt data solution/refinement strategy or even recollect data Computational Crystallography Initiative Physical Biosciences Division

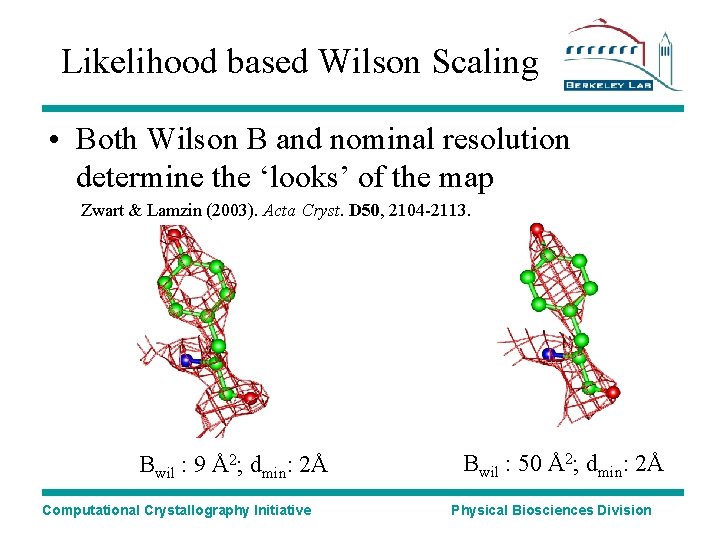
Likelihood based Wilson Scaling • Both Wilson B and nominal resolution determine the ‘looks’ of the map Zwart & Lamzin (2003). Acta Cryst. D 50, 2104 -2113. Bwil : 9 Å2; dmin: 2Å Computational Crystallography Initiative Bwil : 50 Å2; dmin: 2Å Physical Biosciences Division

Likelihood based Wilson Scaling • Data can be anisotropic • Traditional ‘straight line fitting’ not reliable at low resolution • Solution: Likelihood based Wilson scaling – Similar to maximum likelihood refinement, but with absence of knowledge of positional parameters – Results in estimate of anisotropic overall B value. Zwart, Grosse-Kunstleve & Adams, CCP 4 newletter, 2005. Computational Crystallography Initiative Physical Biosciences Division

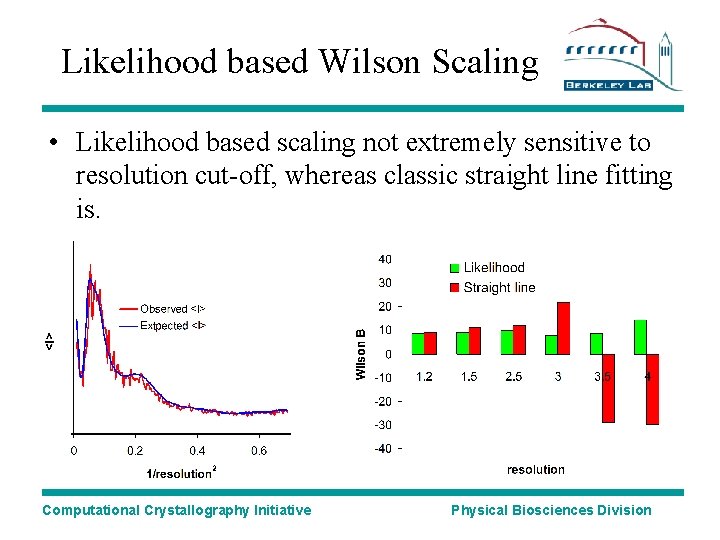
Likelihood based Wilson Scaling • Likelihood based scaling not extremely sensitive to resolution cut-off, whereas classic straight line fitting is. Computational Crystallography Initiative Physical Biosciences Division

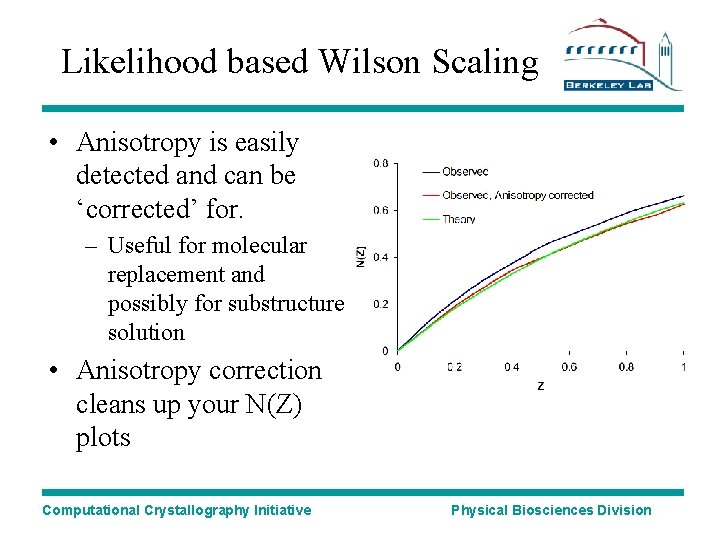
Likelihood based Wilson Scaling • Anisotropy is easily detected and can be ‘corrected’ for. – Useful for molecular replacement and possibly for substructure solution • Anisotropy correction cleans up your N(Z) plots Computational Crystallography Initiative Physical Biosciences Division

Likelihood based Wilson Scaling • Useful by products – For the ML Wilson scaling an ‘expected Wilson plot’ is needed • Using correction term formalism Zwart & Lamzin (2004) Acta Cryst D 60, 220 -226. – Obtained from over 2000 high quality experimental datasets – ‘Expected intensity’ and its standard deviation obtained Computational Crystallography Initiative Physical Biosciences Division

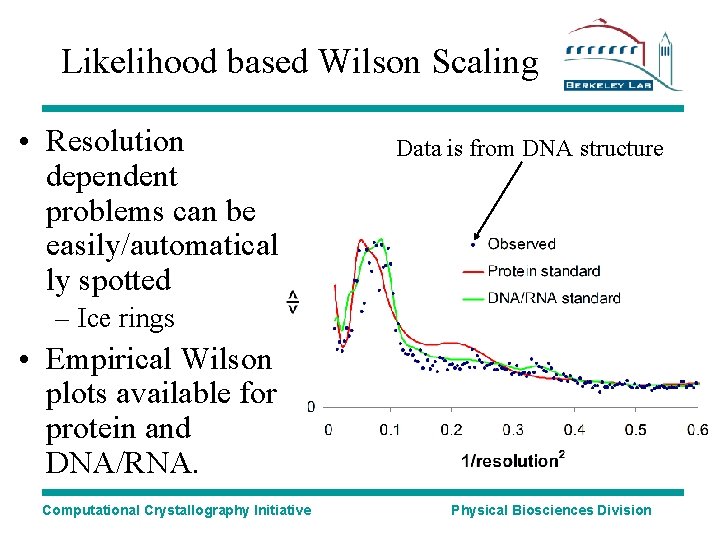
Likelihood based Wilson Scaling • Resolution dependent problems can be easily/automatical ly spotted Data is from DNA structure – Ice rings • Empirical Wilson plots available for protein and DNA/RNA. Computational Crystallography Initiative Physical Biosciences Division

Pseudo Translational Symmetry • Can cause problems in refinement and MR – Incorrect likelihood function due to effects of extra translational symmetry on intensity • Can cause problems or be helpful during MR – Effective ASU is smaller is T-NCS info is used. • The presence of pseudo centering can be detected from an analyses of the Patterson map. – A Fobs Patterson with truncated resolution should reveal a significant off-origin peak. Computational Crystallography Initiative Physical Biosciences Division

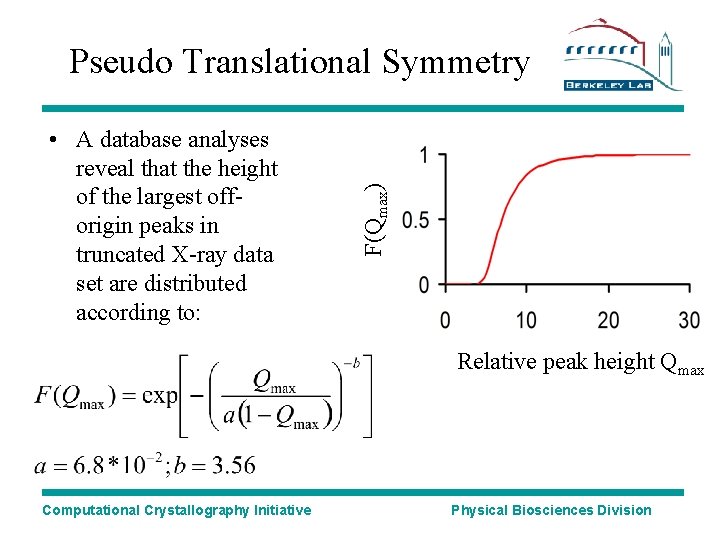
• A database analyses reveal that the height of the largest offorigin peaks in truncated X-ray data set are distributed according to: F(Qmax) Pseudo Translational Symmetry Relative peak height Qmax Computational Crystallography Initiative Physical Biosciences Division

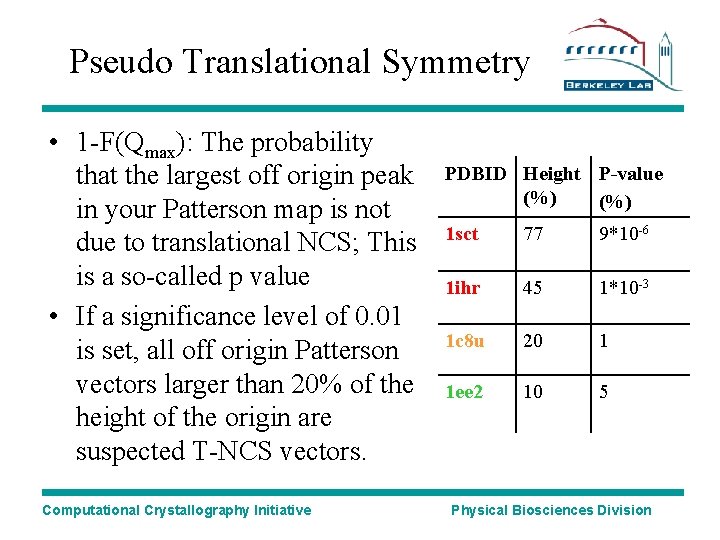
Pseudo Translational Symmetry • 1 -F(Qmax): The probability that the largest off origin peak in your Patterson map is not due to translational NCS; This is a so-called p value • If a significance level of 0. 01 is set, all off origin Patterson vectors larger than 20% of the height of the origin are suspected T-NCS vectors. Computational Crystallography Initiative PDBID Height P-value (%) 1 sct 77 9*10 -6 1 ihr 45 1*10 -3 1 c 8 u 20 1 1 ee 2 10 5 Physical Biosciences Division

Twinning • Merohedral twinning can occur when the lattice has a higher symmetry than the intensities. • When twinning does occur, the recorded intensities are the sum of two independent intensities. – Normal Wilson statistics break down • Detect twinning using intensity statistics Computational Crystallography Initiative Physical Biosciences Division

• Cumulative intensity distribution can be used to identify twinning (acentric data) Pseudo centering Normal Perfect twin Computational Crystallography Initiative N(Z) Twinning Z Physical Biosciences Division

Twinning Pseudo centering + twinning = N(Z) looks normal • Anisotropy in diffraction data produces similar trend to Pseudo centering – Anisotropy can however be removed • How to detect twinning in presence of T-NCS? – Partition miller indices on basis of detected T-NCS vectors • Intensities of subgroups follow normal Wilson statistics (approximately) Computational Crystallography Initiative Physical Biosciences Division

Twinning - 2 + + +; /N Computational Crystallography Initiative 2 <L> Physical Biosciences Division

Twinning • A data base analyses on highly quality, untwinned datasets reveals that the values of the first and second moment of L follow a narrow distribution • This distribution can be used to determine a multivariate Z-score – Large values indicate twinning Computational Crystallography Initiative Physical Biosciences Division

Twinning • Determination of twin laws – From first principles • No twin law will be overlooked • PDB analyses: 36% of structures has at least 1 possible twin law – 50. 9% merohedral; 48. 2% pseudo merohedral; 0. 9% both • 27% of cases with twin laws is suspected to be twinned – 10% of whole PDB(!) • Determination of twin fraction – Fully automated Britton and H analyses as well as ML estimate of twin fraction of basis of L statistic. Computational Crystallography Initiative Physical Biosciences Division

Twinning • Conflicting information – Twin law is present • lattice has higher symmetry than assumed symmetry of intensities – Estimated twin fraction is close to 0. 5 • ‘twin’ related intensities are very similar – <L> test does not indicate twinning • Very strong NCS • Space group too low Computational Crystallography Initiative Physical Biosciences Division

Twinning • Maybe an example of a too low symmetry? Computational Crystallography Initiative Physical Biosciences Division

Anomalous data • Structure solution via experimental methods (especially SAD) is on the rise. • How to identify the presence of anomalous signal? – <DI/I> ; <DF/F> • VERY sensitive to noise – <DI/s. DI>; <DF/s. DF> • 2? – Measurability • Fraction of Bijvoet differences for which – DI/s. DI>3 and (I+/s. I(+) and I(-)/s. I(-) > 3) • Easy to interpret – At 3 Angstrom 6% of Bijvoet pairs are significantly larger than zero Computational Crystallography Initiative Physical Biosciences Division

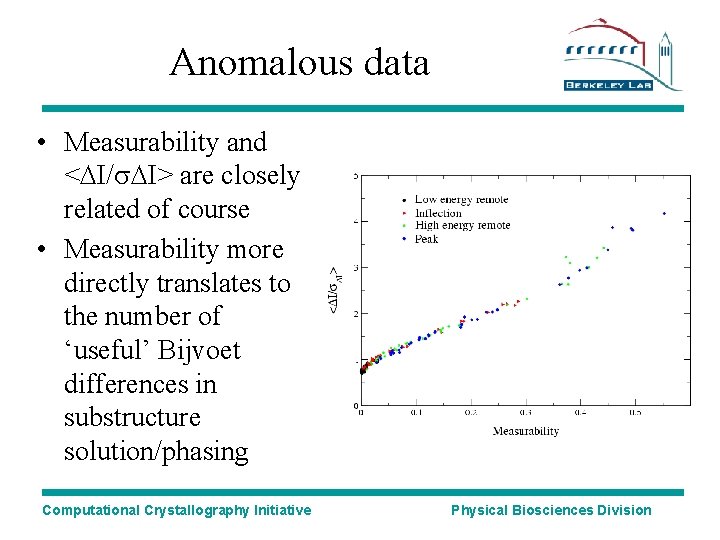
Anomalous data • Measurability and <DI/s. DI> are closely related of course • Measurability more directly translates to the number of ‘useful’ Bijvoet differences in substructure solution/phasing Computational Crystallography Initiative Physical Biosciences Division

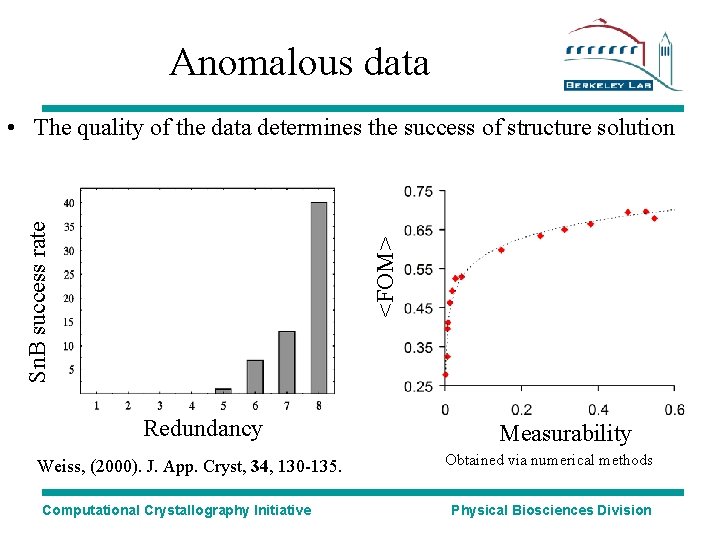
Anomalous data <FOM> Sn. B success rate • The quality of the data determines the success of structure solution Redundancy Weiss, (2000). J. App. Cryst, 34, 130 -135. Computational Crystallography Initiative Measurability Obtained via numerical methods Physical Biosciences Division

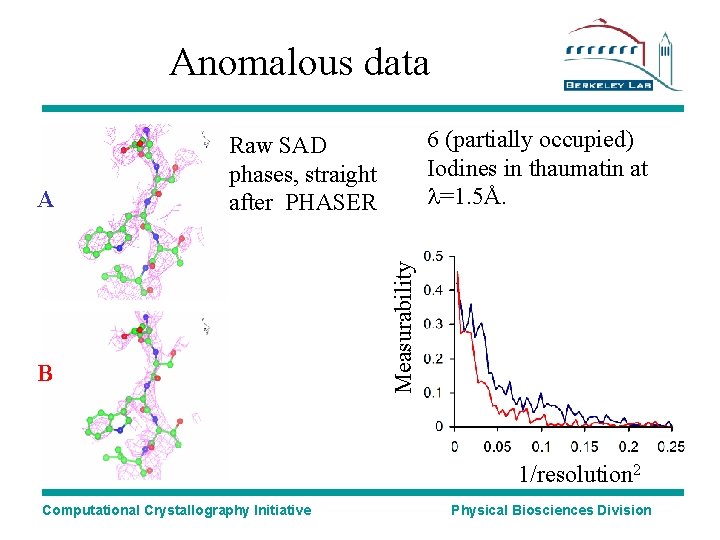
Anomalous data B Measurability A 6 (partially occupied) Iodines in thaumatin at l=1. 5Å. Raw SAD phases, straight after PHASER 1/resolution 2 Computational Crystallography Initiative Physical Biosciences Division

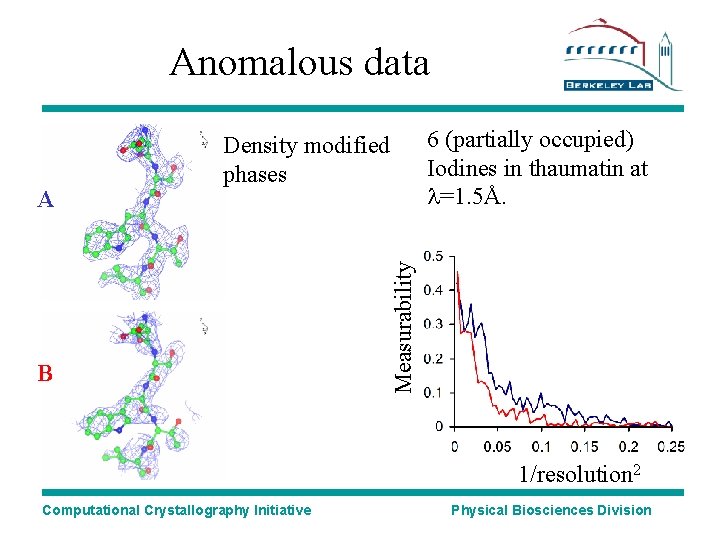
Anomalous data B 6 (partially occupied) Iodines in thaumatin at l=1. 5Å. Measurability A Density modified phases 1/resolution 2 Computational Crystallography Initiative Physical Biosciences Division

Anomalous data • Lys. Os PHASER maps • Ferrodoxin PHASER maps Computational Crystallography Initiative Physical Biosciences Division

Discussion & Conclusions • Software tools are available to point out specific problems – mmtbx. xtriage <input_reflection_file> [params] • Log file are not just numbers, but also contains an extensive interpretation of the statistics • Knowing the idiosyncrasies of your X-ray data might avoid falling in certain pitfalls. – Undetected twinning for instance Computational Crystallography Initiative Physical Biosciences Division

Discussion & Conclusions mmtbx. xtriage at the beamline If problem are detected while at the beamline, possible problems could be solved by recollecting data or adpating the data collection strategy. The Surgeon and the Peasant – 1524. Lucas van Leyden Computational Crystallography Initiative Physical Biosciences Division

Discussion & Conclusions mmtbx. xtriage at home The anatomical lesson of dr. Nicolaes Tulp - 1632. Rembrandt van Rijn. Computational Crystallography Initiative Physical Biosciences Division

Ackowledgements Paul Adams Ralf Grosse-Kunstleve Pavel Afonine Nigel Moriarty Nick Sauter Michael Hohn Cambridge Randy Read Airlie Mc. Coy Laurent Storonoy Los Alamos Tom Terwilliger Li Wei Hung Thirumugan Rhadakanan Texas A&M Univeristy Jim Sachetini Tom Ioerger Eric Mc. Kee Computational Crystallography Initiative Physical Biosciences Division