Finding Sequence Similarities There are many programs used











- Slides: 18

Finding Sequence Similarities There are many programs used to do this. They range from relatively slow programs which find the exact best matching alignment, through ones which take progressively inexact shortcuts to speed things up. Of this latter class, the best known, and easily most widely used is BLAST, developed by Stephen Altschul and others, and continuously refined over the last 10 -15 years. The essential idea is to compare your query sequence against a collection or ‘database’ of target sequences, looking for the one(s) that match the query sequence the best. database query >query AGACGAACCTAGCACAAGCGCGTCTGGAAAGACCCGCCAGCTACGGTCACCGAG CTTCTCATTGCTCTTCCTAACAGTGTGATAGGCTAACCGTAATGGCGTTCAGGA GTATTTGGACTGCAATATTGGCCCTCGTTCAAGGGCGCCTACCATCACCCGACG GTCATGCCGGTCCCCAGCAGCTGCTAATAACTTCGCTACTCAAGTTACCA CGCTAGCAAAACCCACGGCATACCGTTTACCCTTTAAAATCAGCTTCAACCAGC AACGAA COMPARE LIST MATCHES >target 1 AAAACAGGAATATTTACCGGGTAATGATGCATCTCGAGGTACACAATATACCTG GAGAACCGAATTATGAGTTGGCCACCTTAACGAAACCAGCAGAGAAAATCCAACAT GGCAACACCCCTCTGACTACACTAGAAGGAACTACTATGTAAGAAAACAGCCTGTCCCTT GCAGTTTGAATGACTGGGTGATGCGAAATGGGGGTCCTGCCATAGAGCGCTTCCATGGTT TACCTTGCACATTTCAGAGAAGTCCTATGCCAGGAGTCCTACAGGGCCTTCCTGAA ACTATATATGTGCTTATTCTTGTTTGATTTGGCTTTGCAG >target 2 CTCTTAATTTCTCTTCCTGCAGCTCCCTCGCTTTTTCCCTGTTACATTCAT CTGACTTGAAGAGTTGCAAATTTTCAGTGTTTCTGTTTTTGTTGCTGATATGTTGTAAAC TTTTTAATAAAATCTATTTCTATAG >target 3 GCAGTTTGAATGACTGGGTGATGCGAAATGGGGGTCCTGCCATAGAGCGCTTCCATGGTT TACCTTGCACATTTCAGAGAAGTCCTATGCCAGGAGTCCTACAGGGCCTTCCTGAA ACTATATATGTGCTTATTCTTGTTTGATTTGGCTTTGCAGCTAGGGTTTTCACCTTTTCT GGAAAAATACTGGCTTCC >target 4 CTGCTATTAATGGGCAAAACAACTCAAATAAAGTCCCTCTGCCACCCTCAGACACTGCCC CTGGCCCCCAGCTGCCCGCTGATCCTTGTAGCCAGAGCAGTAAAGTTTTGAAAGTGGAGC CCAAGGAGAATAAAGTTATTAAAGAAACTGGCTTTGAACAAGGTGAAAAGTCTTGTGCAG CACCTCTAGATCATACTGTGAAGGAAAATCTTGGACAAACTTCTAAAGAACAGGTGGTAG

Flavours of BLAST can perform a number of similar tasks with different types of sequence: BLASTn – comparing nucleotide sequence vs. nucleotide sequence database BLASTp – comparing protein sequence vs. protein sequence database BLASTx – comparing nucleotide sequence vs. protein sequence database by translating the nucleotide sequence in all possible reading frames t. BLASTn – comparing protein sequence vs. nucleotide sequence database translated into all possible reading frames t. BLASTx – comparing nucleotide sequence vs. nucleotide sequence database translating both into all possible reading frames The amino acid sequence based programs use a substitution matrix to allow some amino acids to count as effective matches with each other. These are the BLOSUM and PAM matrices you may see referred to from time to time.

How does it work? The main task of any sequence comparison program is to test all possible mutual alignments of two sequence and see how good the match is: query 1 st database sequence CCGAGCTTCTCATTGCTCTTCCTAACAGTGTGATAGGCTAACCGTAATGGCGTTC CCGAGCTTCTCATTGCTCTTCCTAACAGTG =TGATAGGCTAACCGTAATGGCGTTC |||||||||||||||||||||||| | ||||||||||||| || || || CTTCTCATTGCTCTTCCTAACAGTGATGATAGGCTAACCGTAATGGCGTTCAGGAGT BLAST achieves its speed through two strategies: It ‘indexes’ the database sequences so it know where all the minor subsequences are in each sequence, so it doesn’t have to look all the way through each sequence each time, letter by letter. It’s ‘word based’, so that it will only start looking for possible extensive alignments once it’s found a seed alignment of an exact match. The default seed lengths are 11 letters for BLASTn and 3 for BLASTp. This means that some good alignments are un-findable, e. g. a protein match with exactly every second amino acid matching. It relies on these ‘uniformly distributed’ alignments being very rare occurrences.

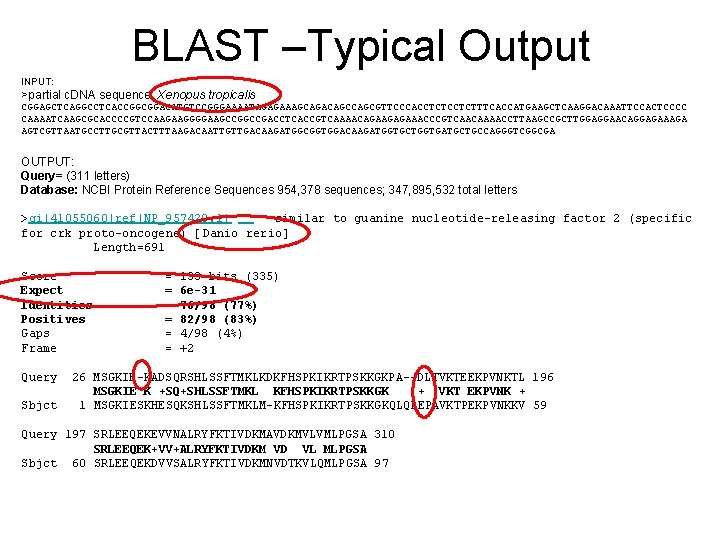
BLAST –Typical Output INPUT: >partial c. DNA sequence, Xenopus tropicalis CGGAGCTCAGGCCTCACCGGCGGACATGTCCGGGAAAATAGAGAAAGCAGACAGCGTTCCCACCTCTTTCACCATGAAGCTCAAGGACAAATTCCACTCCCC CAAAATCAAGCGCACCCCGTCCAAGAAGGGGAAGCCGACCTCACCGTCAAAACAGAAGAGAAACCCGTCAACAAAACCTTAAGCCGCTTGGAGGAACAGGAGAAAGA AGTCGTTAATGCCTTGCGTTACTTTAAGACAATTGTTGACAAGATGGCGGTGGACAAGATGGTGCTGGTGATGCTGCCAGGGTCGGCGA OUTPUT: Query= (311 letters) Database: NCBI Protein Reference Sequences 954, 378 sequences; 347, 895, 532 total letters >gi|41055060|ref|NP_957420. 1| similar to guanine nucleotide-releasing factor 2 (specific for crk proto-oncogene) [Danio rerio] Length=691 Score Expect Identities Positives Gaps Frame = 133 bits (335) = 6 e-31 = 76/98 (77%) = 82/98 (83%) = 4/98 (4%) = +2 Query 26 MSGKIE-KADSQRSHLSSFTMKLKDKFHSPKIKRTPSKKGKPA--DLTVKTEEKPVNKTL 196 MSGKIE K +SQ+SHLSSFTMKL KFHSPKIKRTPSKKGK + VKT EKPVNK + Sbjct 1 MSGKIESKHESQKSHLSSFTMKLM-KFHSPKIKRTPSKKGKQLQPEPAVKTPEKPVNKKV 59 Query 197 SRLEEQEKEVVNALRYFKTIVDKMAVDKMVLVMLPGSA 310 SRLEEQEK+VV+ALRYFKTIVDKM VD VL MLPGSA Sbjct 60 SRLEEQEKDVVSALRYFKTIVDKMNVDTKVLQMLPGSA 97

When is a match significant? Here is a ‘typical’ weak alignment from BLASTp: RF---KISDCQHPCTYSH-NQYMTNHMREC----PYNGAATSIPSWHLIVHPSNGQSVSFPQSDPCQIKMNQNLHLVQMMYDMQTTHV F K S+ + C + + N Y N +C P+ + +W +P + D I N M ++ NFSWKKTSEKETNCQFDYPNDY--NEQTQCQPMTPFKADVFDLWNWEFNANPKLENGIRDLIDDKHDILQIFN------MCWLEVNSS In fact the sequences were randomly generated, so there is no biologically significant alignment… RFKISDCQHPCTYSHNQYMTNHMRECPYNGAATSIPSWHLIVHPSNGQSVSFPQSDPCQIKMNQNLHLVQMMYDMQTTHV NFSWKKTSEKETNCQFDYPNDYNEQTQCQPMTPFKADVFDLWNWEFNANPKLENGIRDLIDDKHDILQIFNMCWLEVNSS

E-values The number of matches like the discovered match that I would expect to find by chance. An E-value of 0. 0 implies that I would expect no matches like this to arise by chance, therefore… An E-value of 1 implies I would expect 1 match like this to arise by chance, so if I have a match with such an E-value…

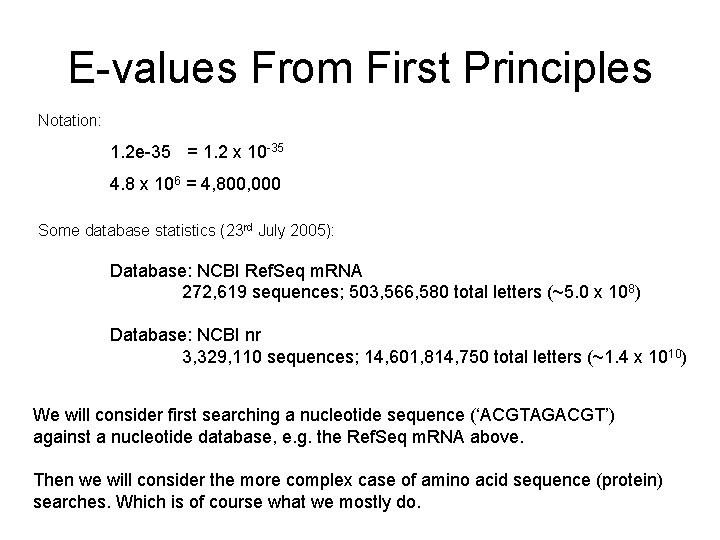
E-values From First Principles Notation: 1. 2 e-35 = 1. 2 x 10 -35 4. 8 x 106 = 4, 800, 000 Some database statistics (23 rd July 2005): Database: NCBI Ref. Seq m. RNA 272, 619 sequences; 503, 566, 580 total letters (~5. 0 x 108) Database: NCBI nr 3, 329, 110 sequences; 14, 601, 814, 750 total letters (~1. 4 x 1010) We will consider first searching a nucleotide sequence (‘ACGTAGACGT’) against a nucleotide database, e. g. the Ref. Seq m. RNA above. Then we will consider the more complex case of amino acid sequence (protein) searches. Which is of course what we mostly do.

Calculating an E-value There are 4 possible nucleotides (ACGT), and the Ref. Seq m. RNA database has ~ 5. 0 x 108 letters in it. The simplest query sequence would be one nucleotide, say ‘A’, so I would, on average, expect ¼ of all the letters in the database to match. Each of these is ‘a match’, so in other words I am expecting (5. 0 x 108) / 4 = ~1. 2 x 108 matches by chance, which is my E-value, 1. 2 x 108. If I increase the query length to two, say ‘AC’, I would expect only ¼ of the ‘A’ matches found in the previous case to be followed by a ‘C’, so my expected number would fall by a factor of four. Now, E-value = ~3 x 107. And so on, for each nucleotide added my expected number of matches, and hence my E-value will fall by a further factor of 4. So for an exact match over 60 nucleotides, this will be (5. 0 x 108) / (4 x 4 x 4 x… 60 times) = (5. 0 x 108) / 1036. so giving an E-value = 5. 0 x 10 -28. We note that this is the sort of value we often see from a BLAST search.

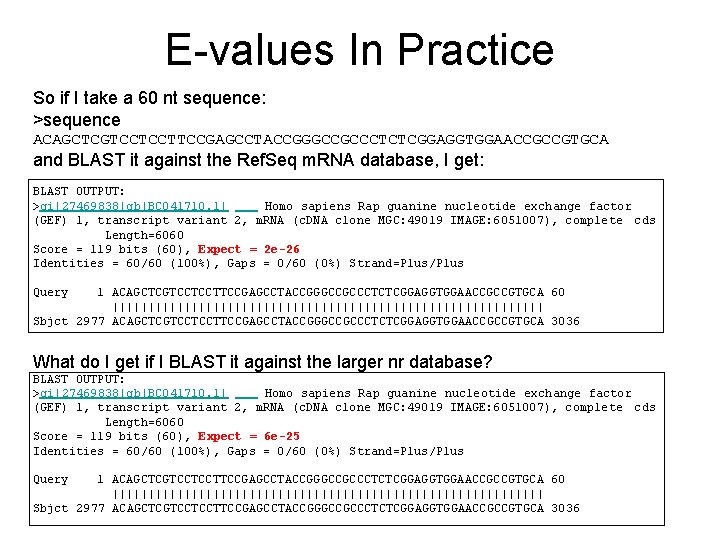
E-values In Practice So if I take a 60 nt sequence: >sequence ACAGCTCGTCCTCCTTCCGAGCCTACCGGGCCGCCCTCTCGGAGGTGGAACCGCCGTGCA and BLAST it against the Ref. Seq m. RNA database, I get: BLAST OUTPUT: >gi|27469838|gb|BC 041710. 1| Homo sapiens Rap guanine nucleotide exchange factor (GEF) 1, transcript variant 2, m. RNA (c. DNA clone MGC: 49019 IMAGE: 6051007), complete cds Length=6060 Score = 119 bits (60), Expect = 2 e-26 Identities = 60/60 (100%), Gaps = 0/60 (0%) Strand=Plus/Plus Query 1 ACAGCTCGTCCTCCTTCCGAGCCTACCGGGCCGCCCTCTCGGAGGTGGAACCGCCGTGCA 60 |||||||||||||||||||||||||||||| Sbjct 2977 ACAGCTCGTCCTCCTTCCGAGCCTACCGGGCCGCCCTCTCGGAGGTGGAACCGCCGTGCA 3036 What do I get if I BLAST it against the larger nr database? BLAST OUTPUT: >gi|27469838|gb|BC 041710. 1| Homo sapiens Rap guanine nucleotide exchange factor (GEF) 1, transcript variant 2, m. RNA (c. DNA clone MGC: 49019 IMAGE: 6051007), complete cds Length=6060 Score = 119 bits (60), Expect = 6 e-25 Identities = 60/60 (100%), Gaps = 0/60 (0%) Strand=Plus/Plus Query 1 ACAGCTCGTCCTCCTTCCGAGCCTACCGGGCCGCCCTCTCGGAGGTGGAACCGCCGTGCA 60 |||||||||||||||||||||||||||||| Sbjct 2977 ACAGCTCGTCCTCCTTCCGAGCCTACCGGGCCGCCCTCTCGGAGGTGGAACCGCCGTGCA 3036

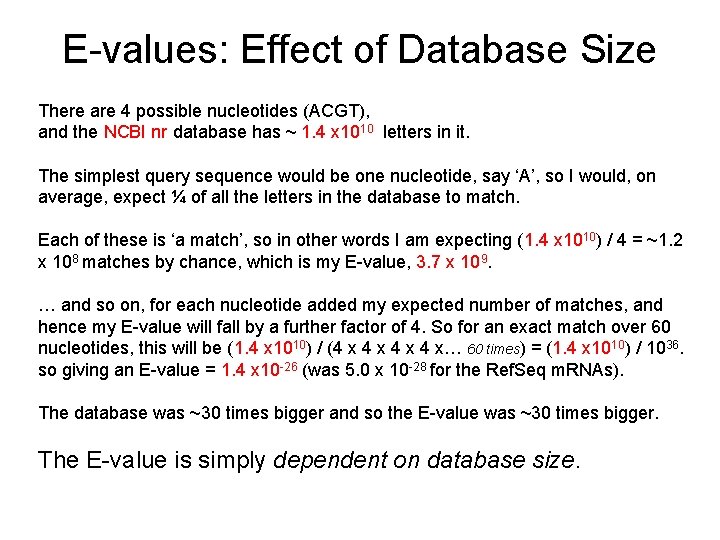
E-values: Effect of Database Size There are 4 possible nucleotides (ACGT), and the NCBI nr database has ~ 1. 4 x 1010 letters in it. The simplest query sequence would be one nucleotide, say ‘A’, so I would, on average, expect ¼ of all the letters in the database to match. Each of these is ‘a match’, so in other words I am expecting (1. 4 x 1010) / 4 = ~1. 2 x 108 matches by chance, which is my E-value, 3. 7 x 109. … and so on, for each nucleotide added my expected number of matches, and hence my E-value will fall by a further factor of 4. So for an exact match over 60 nucleotides, this will be (1. 4 x 1010) / (4 x 4 x 4 x… 60 times) = (1. 4 x 1010) / 1036. so giving an E-value = 1. 4 x 10 -26 (was 5. 0 x 10 -28 for the Ref. Seq m. RNAs). The database was ~30 times bigger and so the E-value was ~30 times bigger. The E-value is simply dependent on database size.

Why were the values different? Our calculated E-value for searching against the Ref. Seq m. RNA database was 5. 0 x 10 -28. But our actual BLAST search at NCBI gave a value of 2. 0 x 10 -26 - about 40 x larger - why is this? Gapped alignments If we were expecting N matches for a query sequence ‘ACGTACGT’, imagine what would happen to N if we allowed gaps in our matches. ACGTAC? GTACGT This would now give us additional possible alignments that would meet our ‘match’ criteria: ACGTACGTACAGTACGT ACGTACGT etc. |||||| |||||| ACGTACGT ACGTAC-GTACGT Hence markedly increasing our E-value, for a given search.

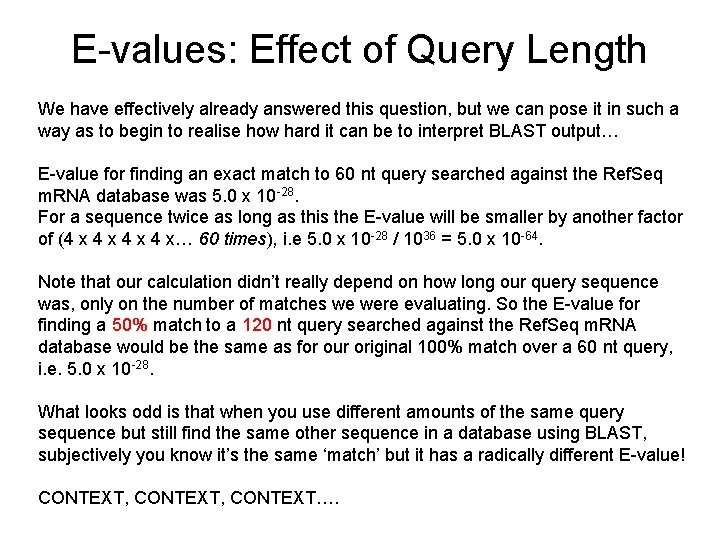
E-values: Effect of Query Length We have effectively already answered this question, but we can pose it in such a way as to begin to realise how hard it can be to interpret BLAST output… E-value for finding an exact match to 60 nt query searched against the Ref. Seq m. RNA database was 5. 0 x 10 -28. For a sequence twice as long as this the E-value will be smaller by another factor of (4 x 4 x 4 x… 60 times), i. e 5. 0 x 10 -28 / 1036 = 5. 0 x 10 -64. Note that our calculation didn’t really depend on how long our query sequence was, only on the number of matches we were evaluating. So the E-value for finding a 50% match to a 120 nt query searched against the Ref. Seq m. RNA database would be the same as for our original 100% match over a 60 nt query, i. e. 5. 0 x 10 -28. What looks odd is that when you use different amounts of the same query sequence but still find the same other sequence in a database using BLAST, subjectively you know it’s the same ‘match’ but it has a radically different E-value! CONTEXT, CONTEXT….

Why not just use % identity? At some levels this a good question. But consider two very different searches, both of which give a 75% identity match Query 1 was 60 nt long: CGGAGCTCAGGGCTTAACGACTGATATCTCCGCGCATGTCGAGAAACGATACAGCG |||||| || |||| | |||||| | ||||| CGGAGCTCAGGCCTCACCGGCGGACATGTCCGGGAAAATAGAGAAAGCAGACAGCG Which would have an E-value ~ 5. 0 x 10 -19 And, Query 2 only 16 nt long: ACGTACGT ||| |||| || ACGCACCTTCGTAGGT Which would have an E-value ~ 30 And intuitively we feel we would expect to see that sort of number of matches in the database just by chance…

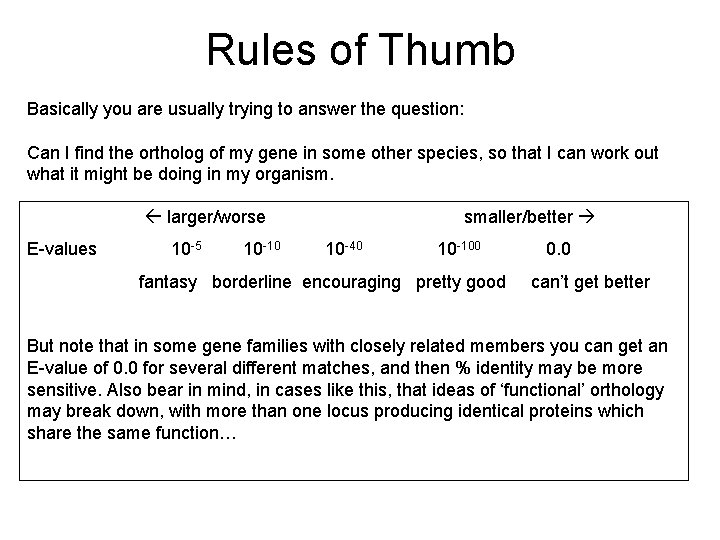
So what’s the real problem? Basically you are usually trying to answer the question: Can I find the ortholog of my gene in some other species, so that I can work out what it might be doing in my organism? And the difficulty is because BLAST does not set out to address questions like orthology. BLAST only tells you about sequence similarity, with some notion of how likely a similarity is to have arisen by chance, based on some general biological principles. You will always have to add in your own knowledge of biology, and exactly what your query sequence was, and how it is related to your matching sequences. In particular whether the degree of similarity matches up to the supposed evolutionary distance between the two species. You will also need to take into account the length of the reported match, compared to the lengths of your query and matched sequences. And of course the size of the database. Are there any useful guidelines though?

Rules of Thumb Basically you are usually trying to answer the question: Can I find the ortholog of my gene in some other species, so that I can work out what it might be doing in my organism. larger/worse E-values 10 -5 10 -10 smaller/better 10 -40 10 -100 fantasy borderline encouraging pretty good 0. 0 can’t get better But note that in some gene families with closely related members you can get an E-value of 0. 0 for several different matches, and then % identity may be more sensitive. Also bear in mind, in cases like this, that ideas of ‘functional’ orthology may break down, with more than one locus producing identical proteins which share the same function…

Protein BLAST It’s (nearly) always better to make comparisons at the amino acid level between protein sequences than the DNA level, because there are many different DNA sequences that can give exactly the same protein sequence. Does this cause us to treat expected values any differently? If we follow the argument as before then for an exact match of a 20 amino acid sequence in the Ref. Seq protein database, each additional amino acid will reduce the E-value by 1/20 th (there are 20 different amino acids). And as there are 347, 895, 532 letters in that database, E-value = ~3. 5 x 108 / (20 x 20 … 20 times) = ~3. 5 x 10 -18. But this is what we get of we run the blast at NCBI: Score = 43. 1 bits (100), Expect = 8 e-04 Identities = 20/20 (100%), Positives = 20/20 (100%), Gaps = 0/20 (0%) Frame = +3 Query 3 SSSSFRAYRAALSEVEPPCI 62 SSSSFRAYRAALSEVEPPCI Sbjct 972 SSSSFRAYRAALSEVEPPCI 991 Really too big a discrepancy to easily explain with hand waving…

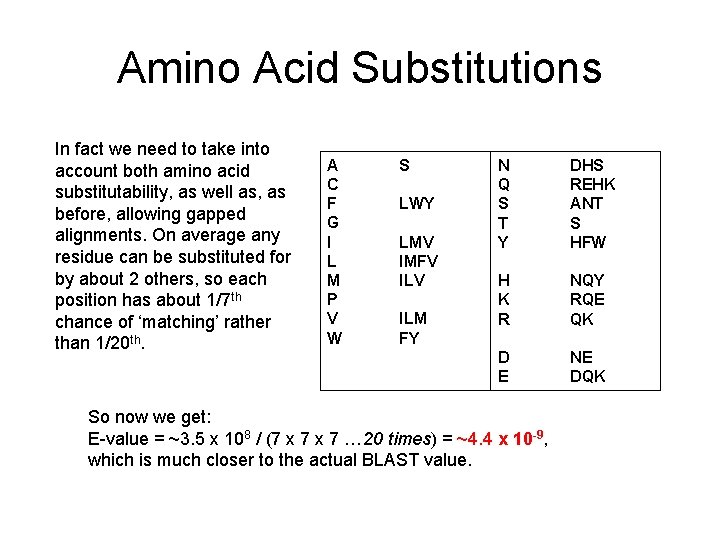
Amino Acid Substitutions In fact we need to take into account both amino acid substitutability, as well as, as before, allowing gapped alignments. On average any residue can be substituted for by about 2 others, so each position has about 1/7 th chance of ‘matching’ rather than 1/20 th. A C F G I L M P V W S LWY LMV IMFV ILM FY N Q S T Y DHS REHK ANT S HFW H K R NQY RQE QK D E NE DQK So now we get: E-value = ~3. 5 x 108 / (7 x 7 … 20 times) = ~4. 4 x 10 -9, which is much closer to the actual BLAST value.

Exercises Go to the file random-DNA-sequences. html in the workshop directory, randomly select one of the 20 randomly generated nucleotide sequences, and do a BLASTx (DNA->protein) search of NCBI nr protein database. Repeat with a few other sequences. Did you find any ‘significant’ hits? What conclusions might you draw from this exercise? Try the same sequence(s) against the nr nucleotide database. Is there any general difference?