Figure 20 2 Bacterium Cell containing gene of
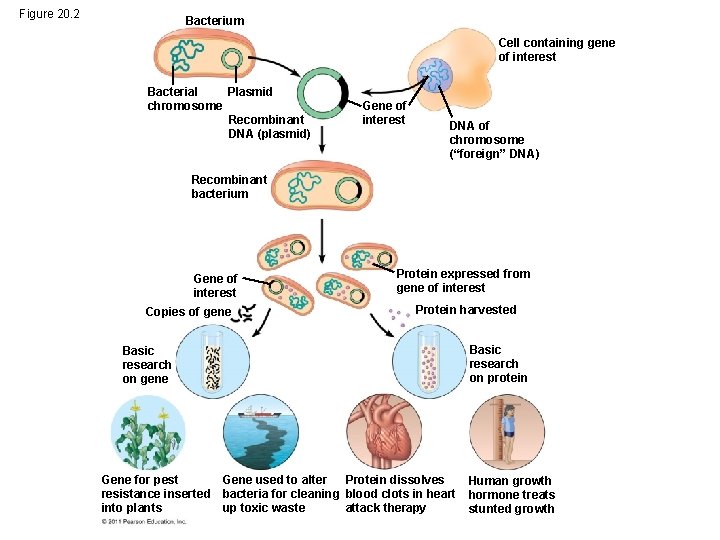
Figure 20. 2 Bacterium Cell containing gene of interest Bacterial Plasmid chromosome Recombinant DNA (plasmid) Gene of interest DNA of chromosome (“foreign” DNA) Recombinant bacterium Gene of interest Copies of gene Protein expressed from gene of interest Protein harvested Basic research on gene Gene for pest Gene used to alter Protein dissolves resistance inserted bacteria for cleaning blood clots in heart into plants up toxic waste attack therapy Basic research on protein Human growth hormone treats stunted growth
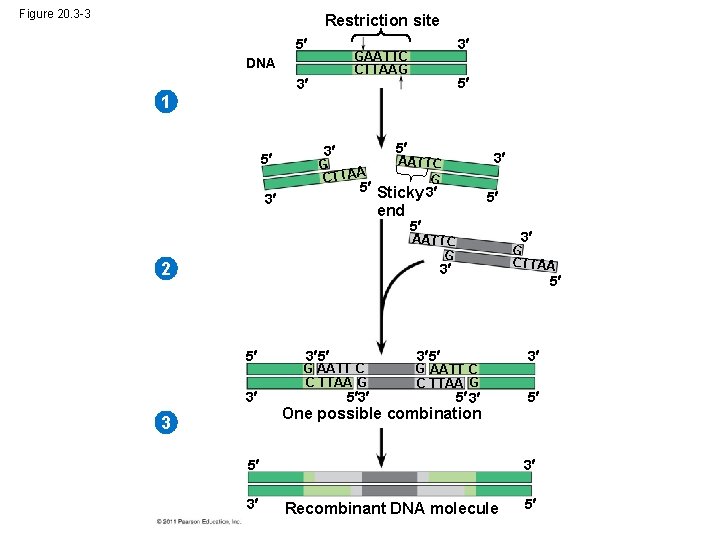
Figure 20. 3 -3 Restriction site 5 3 GAATTC CTTAAG DNA 3 5 1 5 5 3 G CTTAA 3 AATTC G 5 Sticky 3 3 end 5 5 3 AATTC G 2 G CTTAA 3 5 3 3 5 G AATT C C TTAA G 5 3 5 3 5 G AATT C C TTAA G 5 3 3 5 One possible combination 3 5 3 3 Recombinant DNA molecule 5
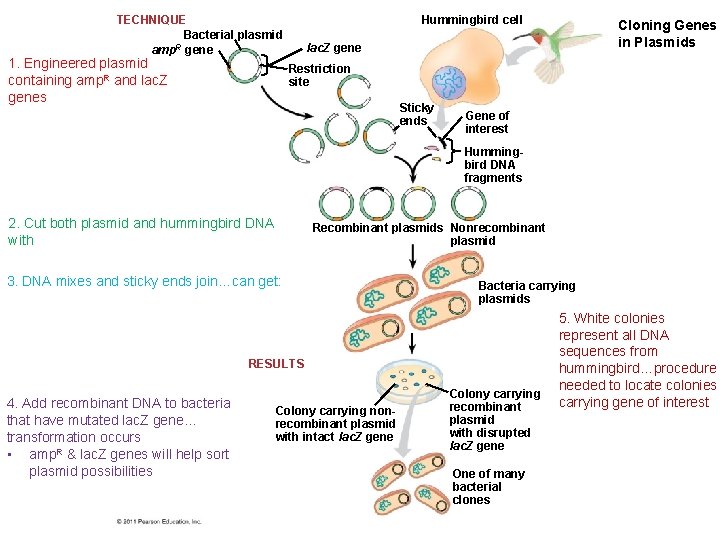
TECHNIQUE Bacterial plasmid R amp gene 1. Engineered plasmid containing amp. R and lac. Z genes Hummingbird cell Cloning Genes in Plasmids lac. Z gene Restriction site Sticky ends Gene of interest Hummingbird DNA fragments 2. Cut both plasmid and hummingbird DNA with Recombinant plasmids Nonrecombinant plasmid 3. DNA mixes and sticky ends join…can get: Bacteria carrying plasmids RESULTS 4. Add recombinant DNA to bacteria that have mutated lac. Z gene… transformation occurs • amp. R & lac. Z genes will help sort plasmid possibilities Colony carrying nonrecombinant plasmid with intact lac. Z gene Colony carrying recombinant plasmid with disrupted lac. Z gene One of many bacterial clones 5. White colonies represent all DNA sequences from hummingbird…procedure needed to locate colonies carrying gene of interest
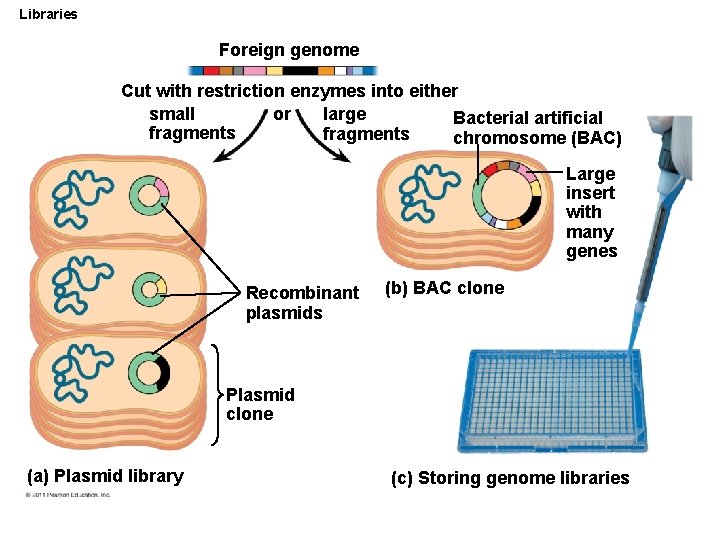
Libraries Foreign genome Cut with restriction enzymes into either small large or Bacterial artificial fragments chromosome (BAC) Large insert with many genes Recombinant plasmids (b) BAC clone Plasmid clone (a) Plasmid library (c) Storing genome libraries
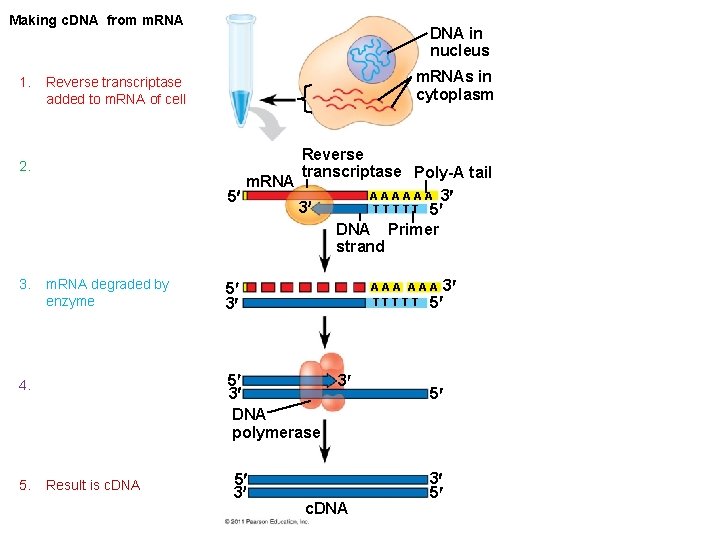
Making c. DNA from m. RNA DNA in nucleus m. RNAs in cytoplasm 1. Reverse transcriptase added to m. RNA of cell 2. a 3. m. RNA degraded by enzyme 5 3 4. A 5 3 DNA polymerase 5. Result is c. DNA Reverse transcriptase Poly-A tail m. RNA A A A 3 5 3 T T T 5 DNA Primer strand 5 3 A A A 3 T T T 5 3 c. DNA 5 3 5
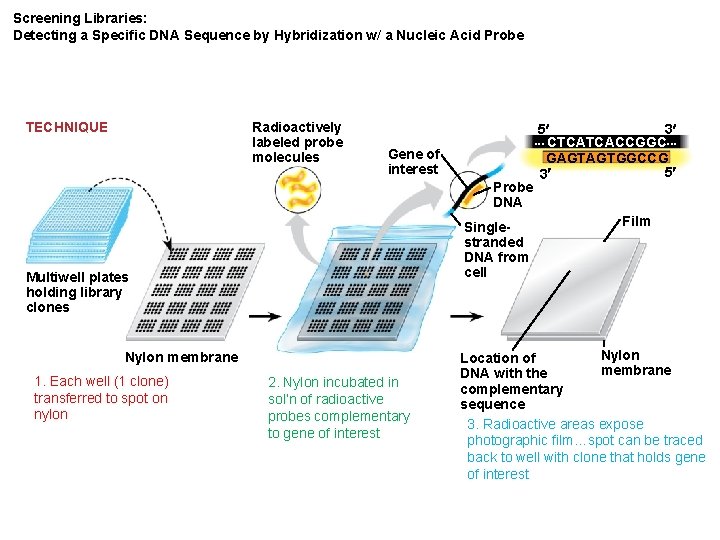
Screening Libraries: Detecting a Specific DNA Sequence by Hybridization w/ a Nucleic Acid Probe Radioactively labeled probe molecules TECHNIQUE Gene of interest Probe DNA Singlestranded DNA from cell Multiwell plates holding library clones Nylon membrane 1. Each well (1 clone) transferred to spot on nylon 2. Nylon incubated in sol’n of radioactive probes complementary to gene of interest 5 3 CTCATCACCGGC GAGTAGTGGCCG 5 3 Film Nylon Location of membrane DNA with the complementary sequence 3. Radioactive areas expose photographic film…spot can be traced back to well with clone that holds gene of interest
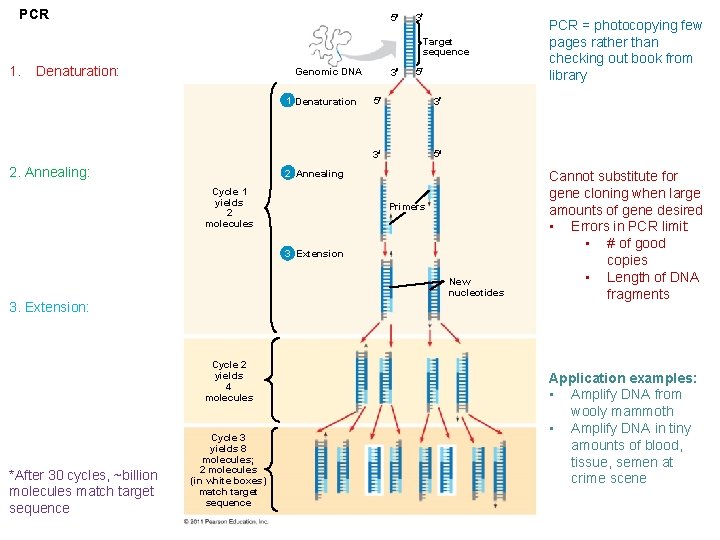
PCR 5 3 Target sequence 1. Denaturation: Genomic DNA 1 Denaturation 2. Annealing: 3 5 5 3 3 5 2 Annealing Cycle 1 yields 2 molecules Primers 3 Extension New nucleotides 3. Extension: Cycle 2 yields 4 molecules *After 30 cycles, ~billion molecules match target sequence Cycle 3 yields 8 molecules; 2 molecules (in white boxes) match target sequence PCR = photocopying few pages rather than checking out book from library Cannot substitute for gene cloning when large amounts of gene desired • Errors in PCR limit: • # of good copies • Length of DNA fragments Application examples: • Amplify DNA from wooly mammoth • Amplify DNA in tiny amounts of blood, tissue, semen at crime scene
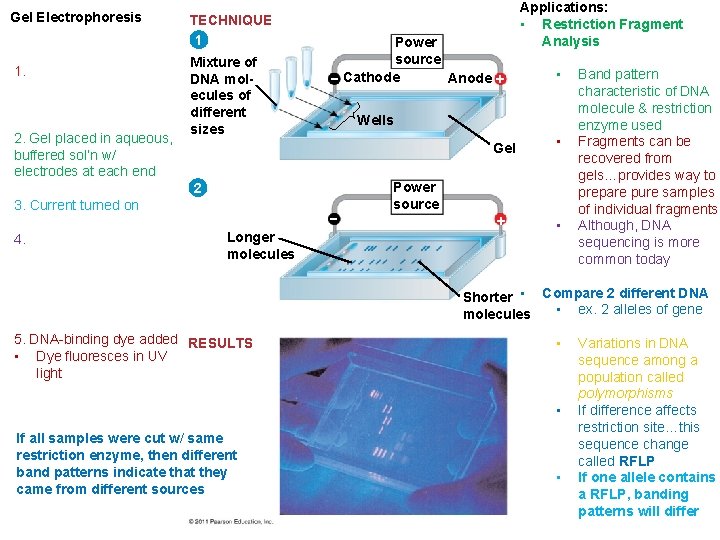
Gel Electrophoresis 1. E 2. Gel placed in aqueous, buffered sol’n w/ electrodes at each end TECHNIQUE 1 Mixture of DNA molecules of different sizes Power source Cathode Anode 4. (-) Longer molecules • Wells 2 3. Current turned on Applications: • Restriction Fragment Analysis Power source Gel • • Band pattern characteristic of DNA molecule & restriction enzyme used Fragments can be recovered from gels…provides way to prepare pure samples of individual fragments Although, DNA sequencing is more common today Shorter • Compare 2 different DNA • ex. 2 alleles of gene molecules 5. DNA-binding dye added RESULTS • Dye fluoresces in UV light • • If all samples were cut w/ same restriction enzyme, then different band patterns indicate that they came from different sources • Variations in DNA sequence among a population called polymorphisms If difference affects restriction site…this sequence change called RFLP If one allele contains a RFLP, banding patterns will differ
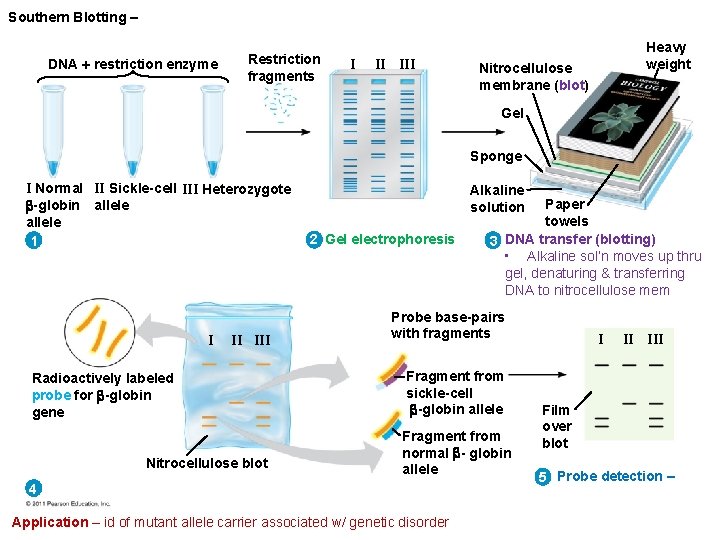
Southern Blotting – DNA restriction enzyme Restriction fragments I II III Heavy weight Nitrocellulose membrane (blot) Gel Sponge I Normal II Sickle-cell III Heterozygote -globin allele 1 I II III Radioactively labeled probe for -globin gene Nitrocellulose blot Alkaline solution 2 Gel electrophoresis Paper towels 3 DNA transfer (blotting) • Alkaline sol’n moves up thru gel, denaturing & transferring DNA to nitrocellulose mem Probe base-pairs with fragments Fragment from sickle-cell -globin allele Fragment from normal - globin allele 4 Application – id of mutant allele carrier associated w/ genetic disorder I II III Film over blot 5 Probe detection –
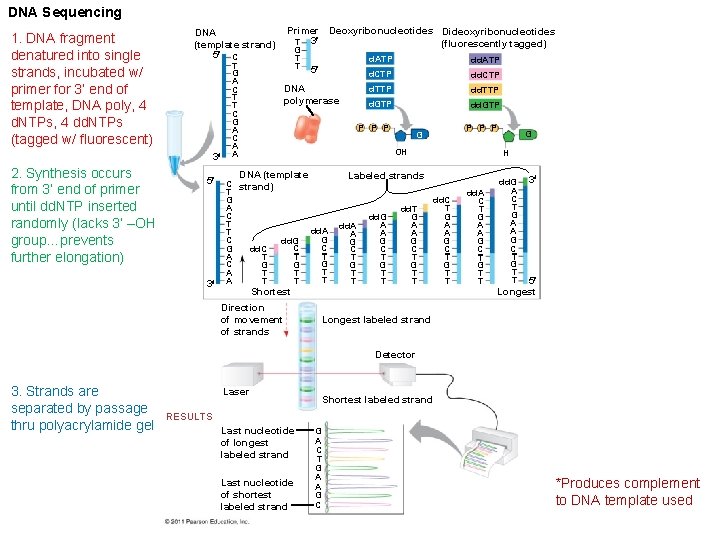
DNA Sequencing 1. DNA fragment denatured into single strands, incubated w/ primer for 3’ end of template, DNA poly, 4 d. NTPs, 4 dd. NTPs (tagged w/ fluorescent) DNA (template strand) 5 C 3 2. Synthesis occurs from 3’ end of primer until dd. NTP inserted randomly (lacks 3’ –OH group…prevents further elongation) 5 3 T G A C T T C G A C A A Primer Deoxyribonucleotides Dideoxyribonucleotides T 3 (fluorescently tagged) G T T 5 DNA polymerase d. ATP d. CTP dd. CTP d. TTP d. GTP dd. GTP P P P G OH DNA (template C strand) T G A C T T C dd. G C G dd. C T A T C G G A T T T A T dd. A G C T G T T Shortest Direction of movement of strands H Labeled strands dd. A A G C T G T T dd. G A A G C T G T T dd. T G A A G C T G T T G dd. C T G A A G C T G T T dd. A C T G A A G C T G T T dd. G A C T G A A G C T G T T 3 5 Longest labeled strand Detector 3. Strands are separated by passage thru polyacrylamide gel Laser Shortest labeled strand RESULTS Last nucleotide of longest labeled strand Last nucleotide of shortest labeled strand G A C T G A A G C *Produces complement to DNA template used
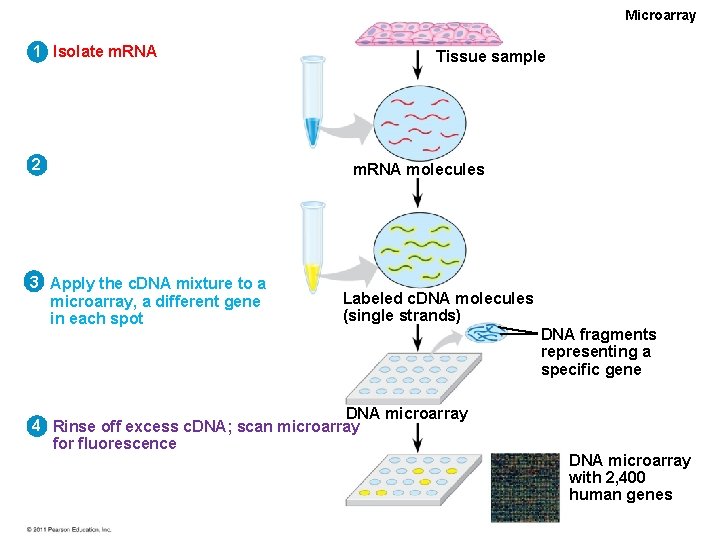
Microarray 1 Isolate m. RNA 2 3 Apply the c. DNA mixture to a microarray, a different gene in each spot Tissue sample m. RNA molecules Labeled c. DNA molecules (single strands) DNA microarray 4 Rinse off excess c. DNA; scan microarray for fluorescence DNA fragments representing a specific gene DNA microarray with 2, 400 human genes
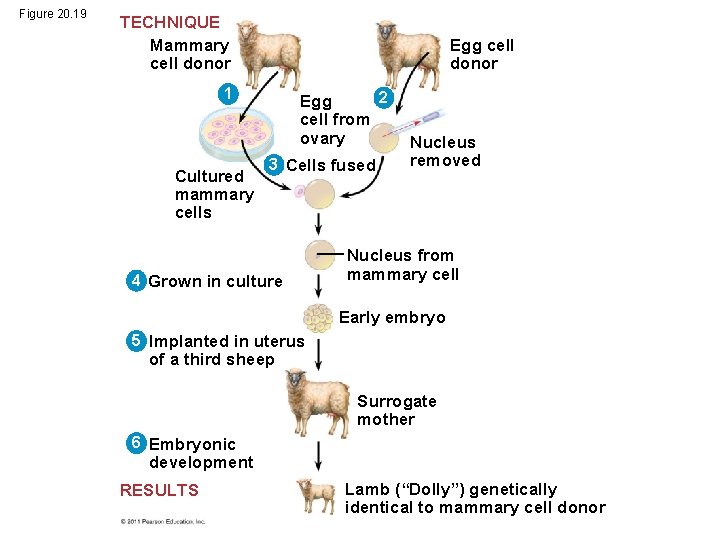
Figure 20. 19 TECHNIQUE Mammary cell donor Egg cell donor 1 Cultured mammary cells 2 Egg cell from ovary 3 Cells fused 4 Grown in culture Nucleus removed Nucleus from mammary cell Early embryo 5 Implanted in uterus of a third sheep Surrogate mother 6 Embryonic development RESULTS Lamb (“Dolly”) genetically identical to mammary cell donor
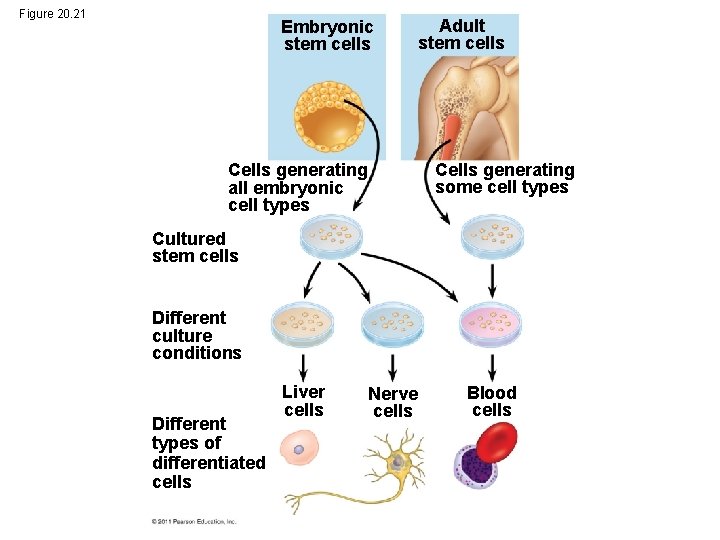
Figure 20. 21 Embryonic stem cells Adult stem cells Cells generating some cell types Cells generating all embryonic cell types Cultured stem cells Different culture conditions Different types of differentiated cells Liver cells Nerve cells Blood cells
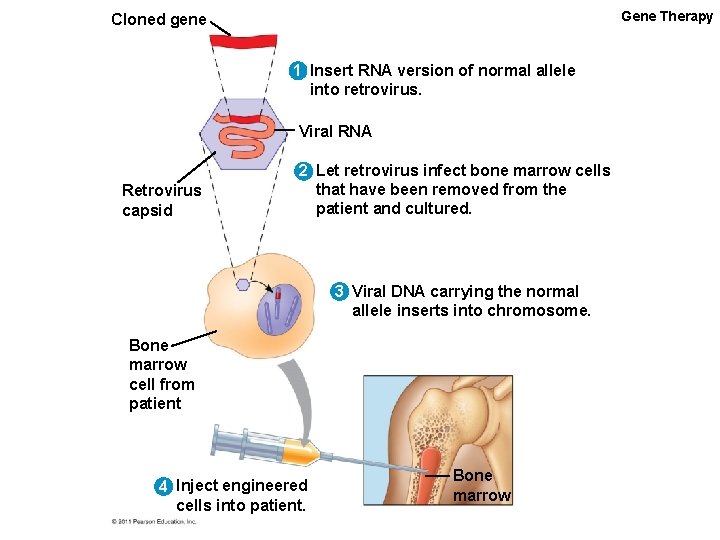
Gene Therapy Cloned gene 1 Insert RNA version of normal allele into retrovirus. Viral RNA Retrovirus capsid 2 Let retrovirus infect bone marrow cells that have been removed from the patient and cultured. 3 Viral DNA carrying the normal allele inserts into chromosome. Bone marrow cell from patient 4 Inject engineered cells into patient. Bone marrow
- Slides: 14