Figure 16 2 EXPERIMENT Living S cells control




- Slides: 37

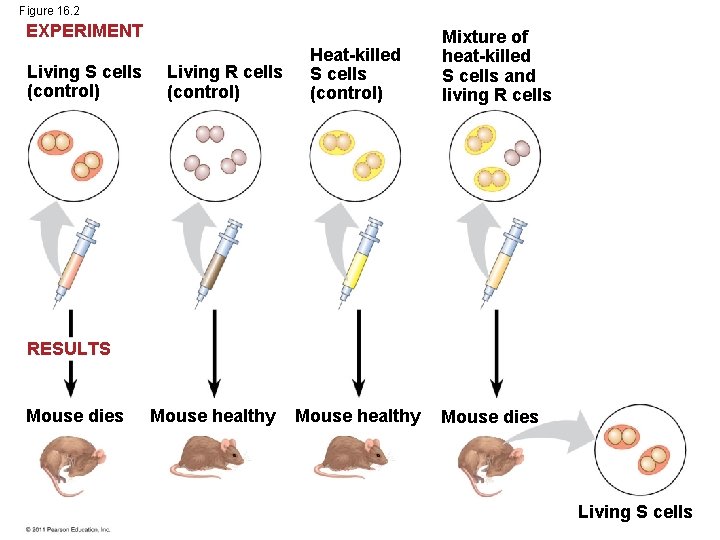
Figure 16. 2 EXPERIMENT Living S cells (control) Living R cells (control) Heat-killed S cells (control) Mixture of heat-killed S cells and living R cells RESULTS Mouse dies Mouse healthy Mouse dies Living S cells

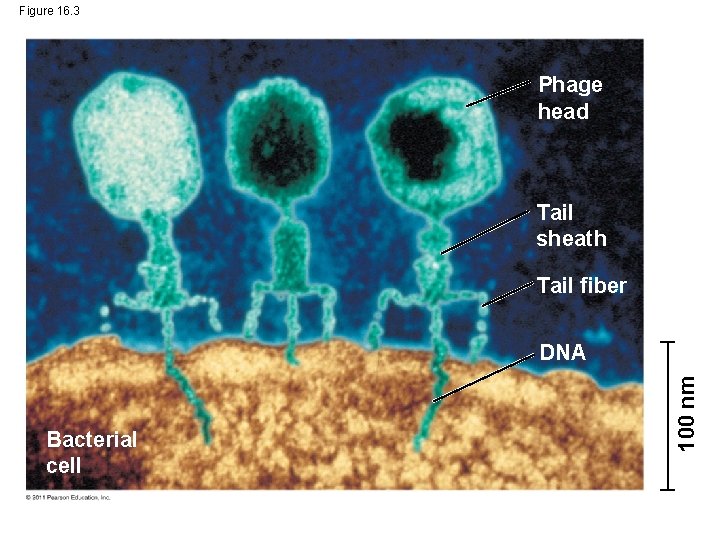
Figure 16. 3 Phage head Tail sheath Tail fiber Bacterial cell 100 nm DNA

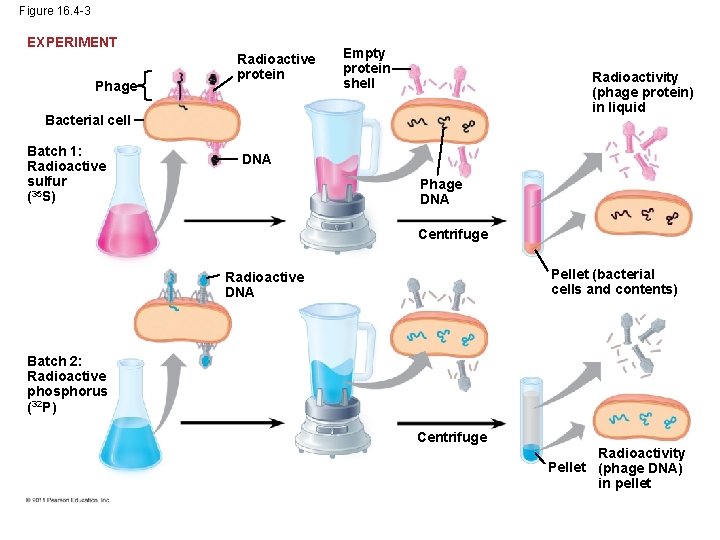
Figure 16. 4 -3 EXPERIMENT Phage Radioactive protein Empty protein shell Radioactivity (phage protein) in liquid Bacterial cell Batch 1: Radioactive sulfur (35 S) DNA Phage DNA Centrifuge Pellet (bacterial cells and contents) Radioactive DNA Batch 2: Radioactive phosphorus (32 P) Centrifuge Radioactivity Pellet (phage DNA) in pellet

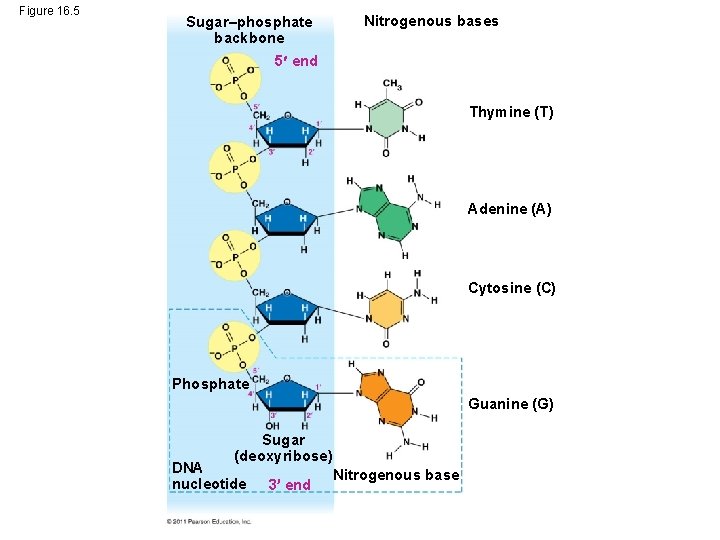
Figure 16. 5 Sugar–phosphate backbone Nitrogenous bases 5 end Thymine (T) Adenine (A) Cytosine (C) Phosphate Guanine (G) Sugar (deoxyribose) DNA nucleotide 3 end Nitrogenous base

Figure 16. 6 (a) Rosalind Franklin (b) Franklin’s X-ray diffraction photograph of DNA

Figure 16. 1

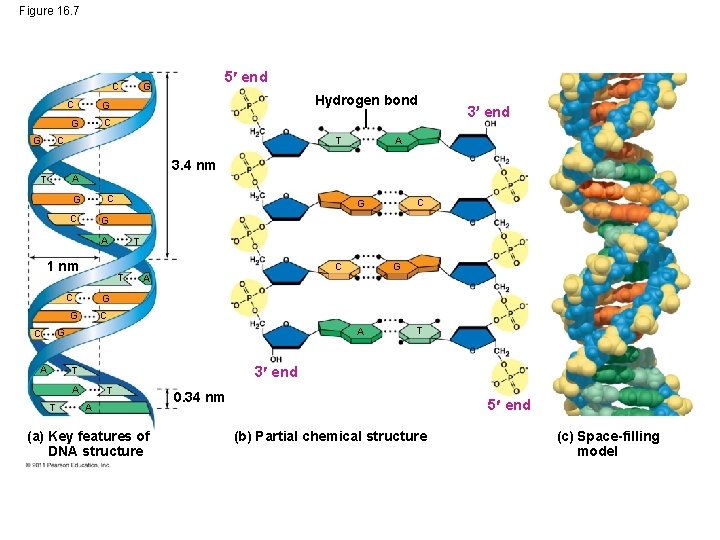
Figure 16. 7 C 5 end G C Hydrogen bond G C G 3. 4 nm C G 1 nm T T C C A G T 3 end T A T G C A G G A C A T 3 end T A (a) Key features of DNA structure 0. 34 nm 5 end (b) Partial chemical structure (c) Space-filling model

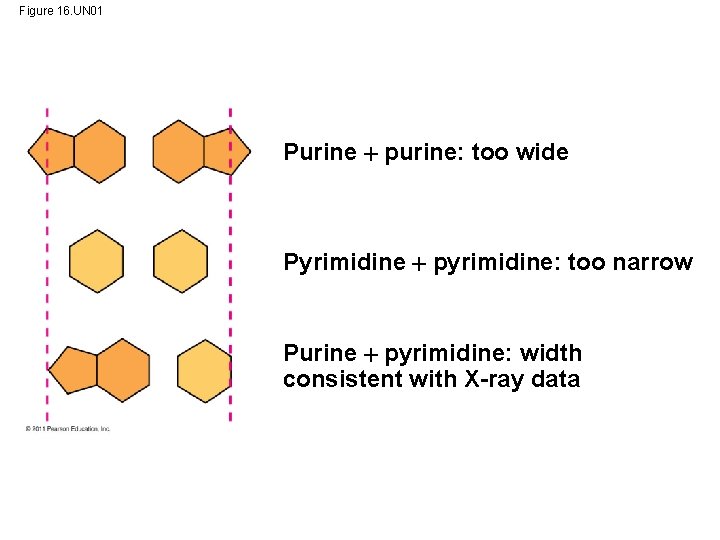
Figure 16. UN 01 Purine purine: too wide Pyrimidine pyrimidine: too narrow Purine pyrimidine: width consistent with X-ray data

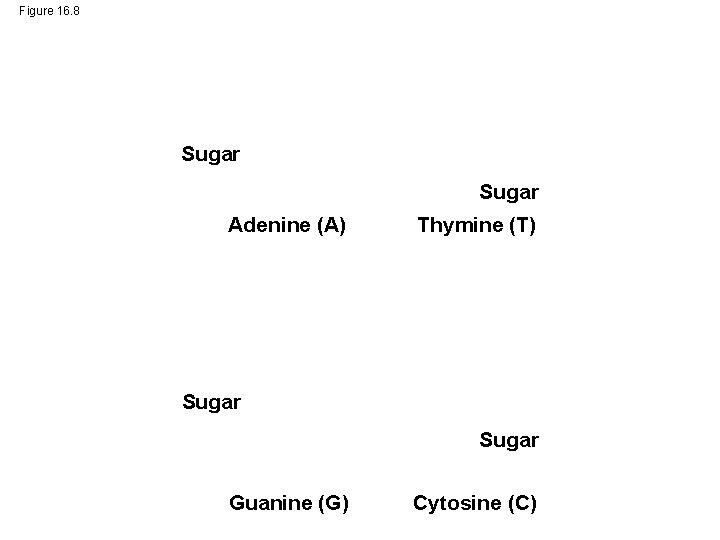
Figure 16. 8 Sugar Adenine (A) Thymine (T) Sugar Guanine (G) Cytosine (C)

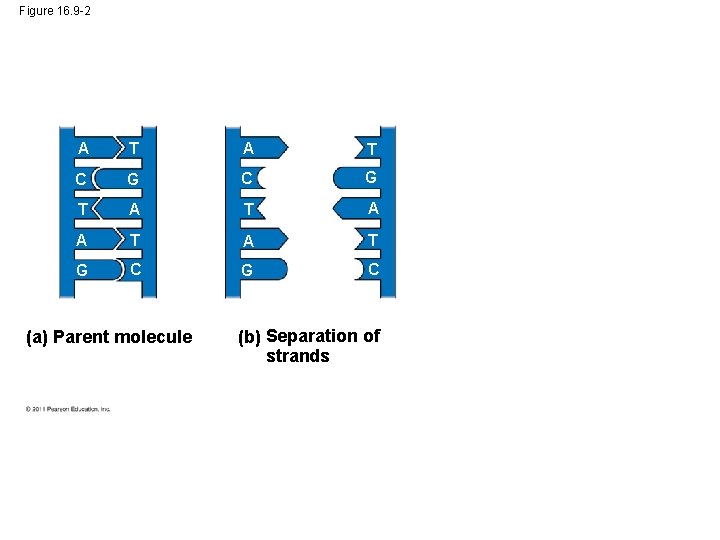
Figure 16. 9 -1 A T C G T A A T G C (a) Parent molecule

Figure 16. 9 -2 A T C G T A A T G C (a) Parent molecule (b) Separation of strands

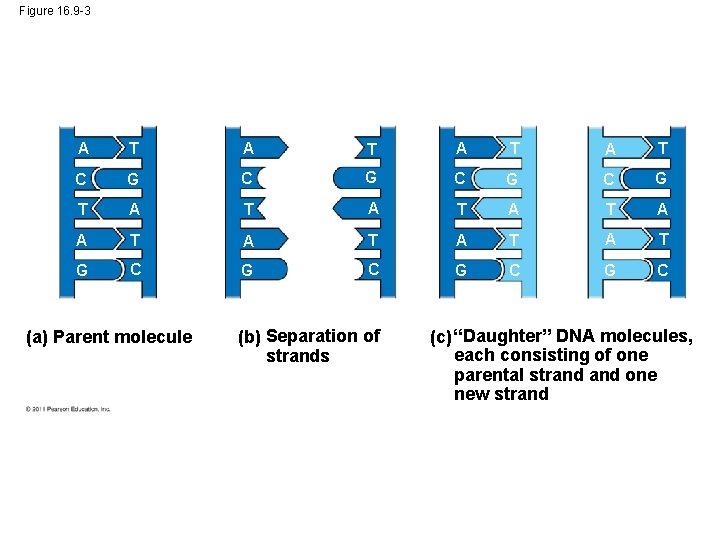
Figure 16. 9 -3 A T A T C G C G T A T A T G C G C (a) Parent molecule (b) Separation of strands (c) “Daughter” DNA molecules, each consisting of one parental strand one new strand

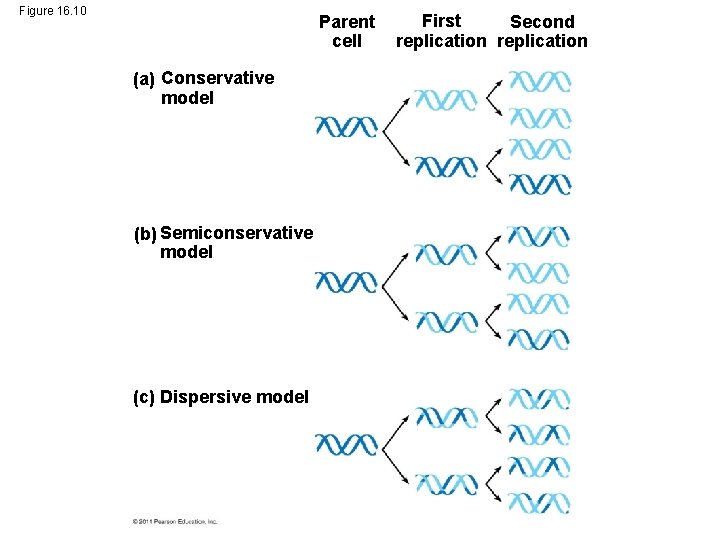
Figure 16. 10 Parent cell (a) Conservative model (b) Semiconservative model (c) Dispersive model First Second replication

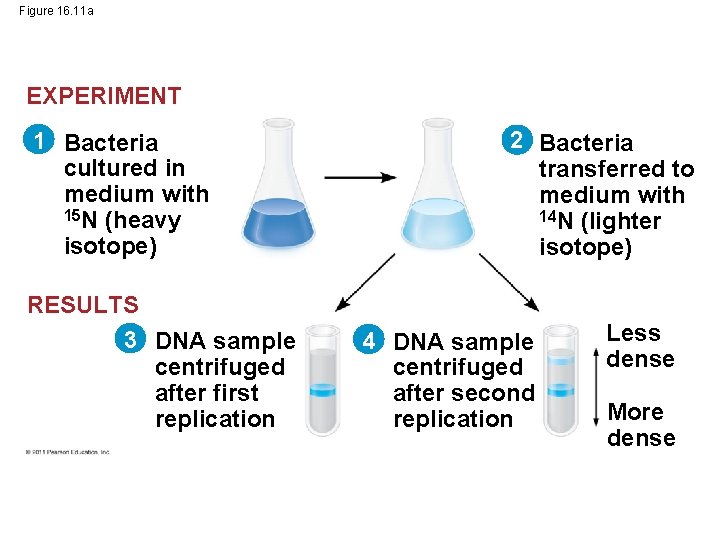
Figure 16. 11 a EXPERIMENT 1 Bacteria cultured in medium with 15 N (heavy isotope) RESULTS 3 DNA sample centrifuged after first replication 2 Bacteria transferred to medium with 14 N (lighter isotope) 4 DNA sample centrifuged after second replication Less dense More dense

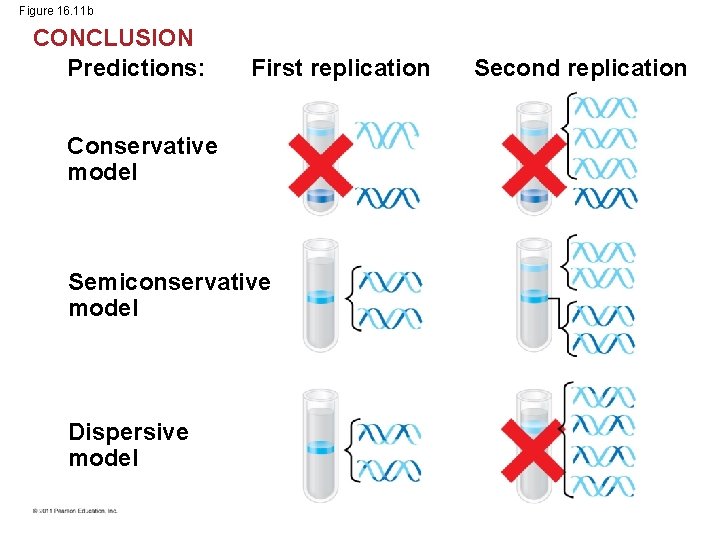
Figure 16. 11 b CONCLUSION Predictions: First replication Conservative model Semiconservative model Dispersive model Second replication

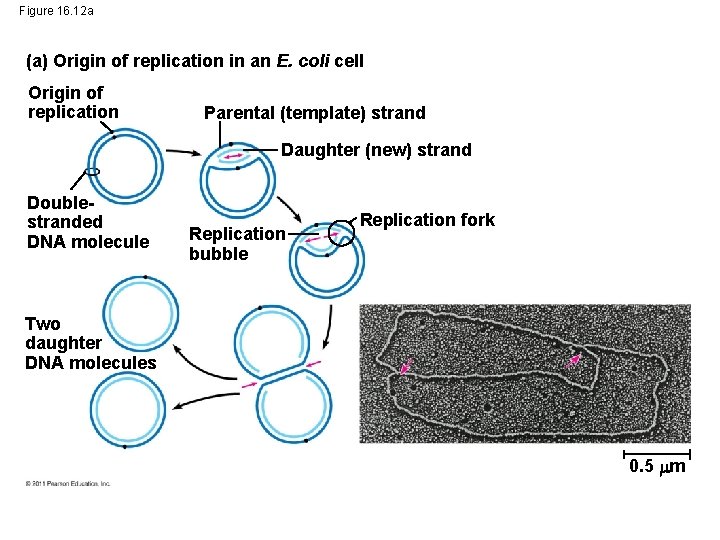
Figure 16. 12 a (a) Origin of replication in an E. coli cell Origin of replication Parental (template) strand Daughter (new) strand Doublestranded DNA molecule Replication bubble Replication fork Two daughter DNA molecules 0. 5 m

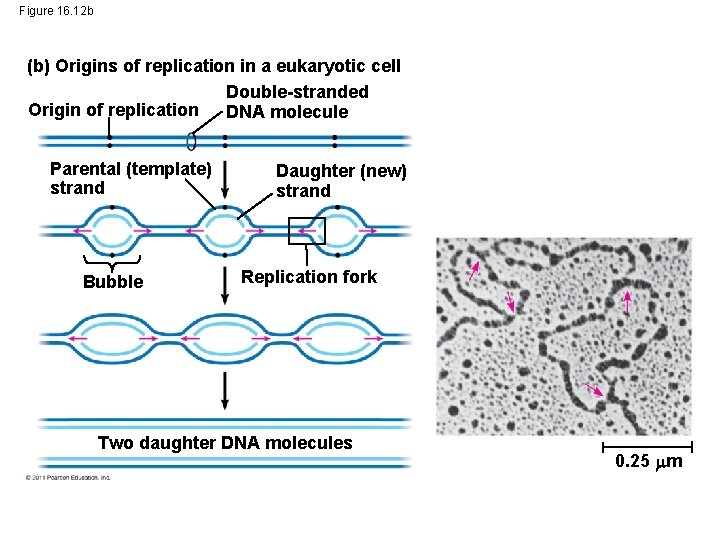
Figure 16. 12 b (b) Origins of replication in a eukaryotic cell Double-stranded Origin of replication DNA molecule Parental (template) strand Bubble Daughter (new) strand Replication fork Two daughter DNA molecules 0. 25 m

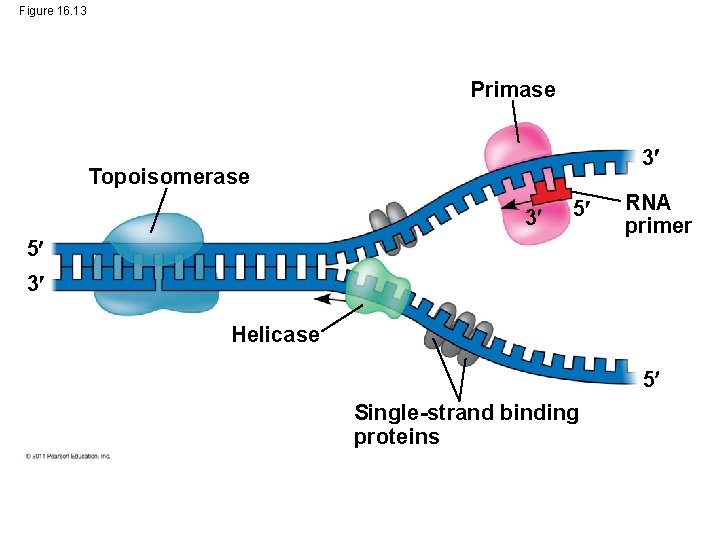
Figure 16. 13 Primase 3 Topoisomerase 3 5 5 RNA primer 3 Helicase 5 Single-strand binding proteins

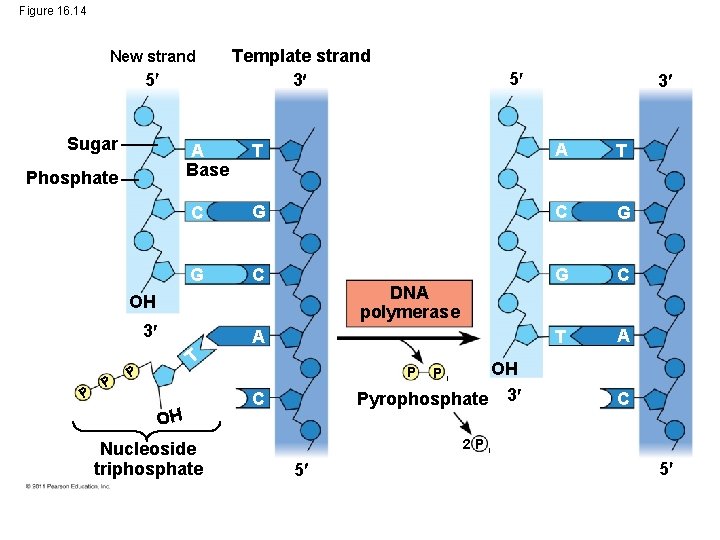
Figure 16. 14 New strand 5 Sugar Phosphate Template strand 3 T A T C G G C T A 3 DNA polymerase P A T P 3 A Base OH P 5 OH Nucleoside triphosphate OH Pyrophosphate 3 P C Pi C 2 Pi 5 5

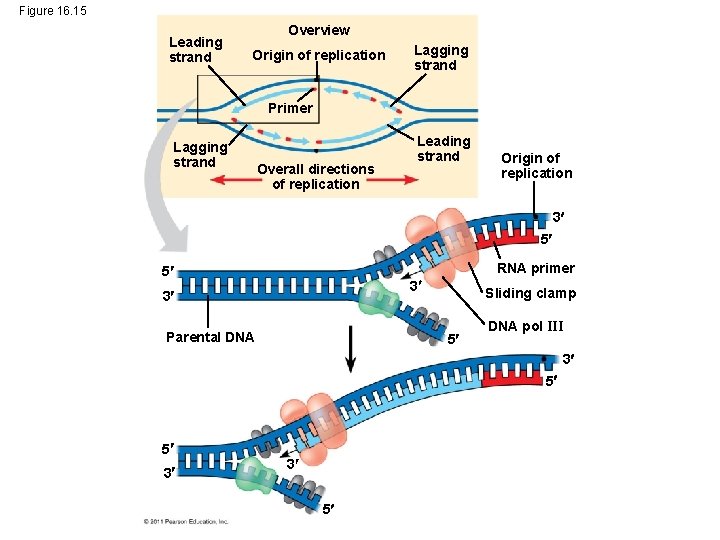
Figure 16. 15 Leading strand Overview Origin of replication Lagging strand Primer Lagging strand Overall directions of replication Leading strand Origin of replication 3 5 RNA primer 5 3 3 Parental DNA Sliding clamp 5 DNA pol III 3 5 5 3 3 5

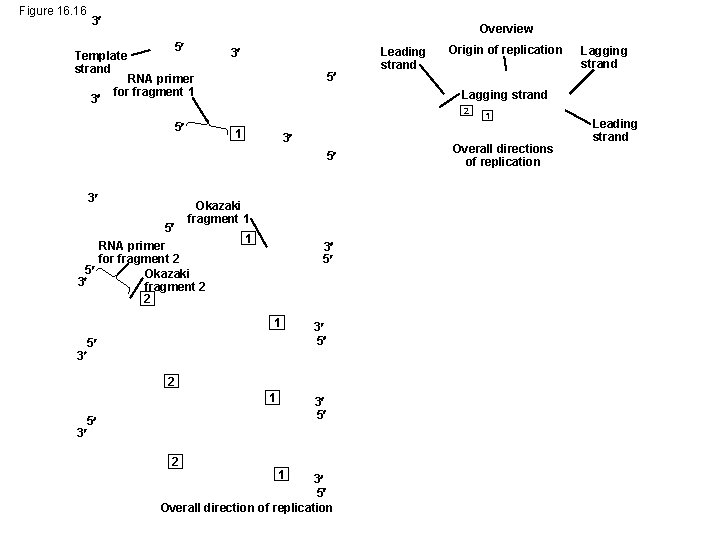
Figure 16. 16 3 Overview 5 Template strand 3 3 5 RNA primer for fragment 1 Leading strand Origin of replication Lagging strand 2 5 1 3 5 Okazaki fragment 1 RNA primer for fragment 2 5 Okazaki 3 fragment 2 2 1 3 5 1 3 5 3 5 2 1 3 3 5 5 2 Lagging strand 1 3 5 Overall direction of replication 1 Overall directions of replication Leading strand

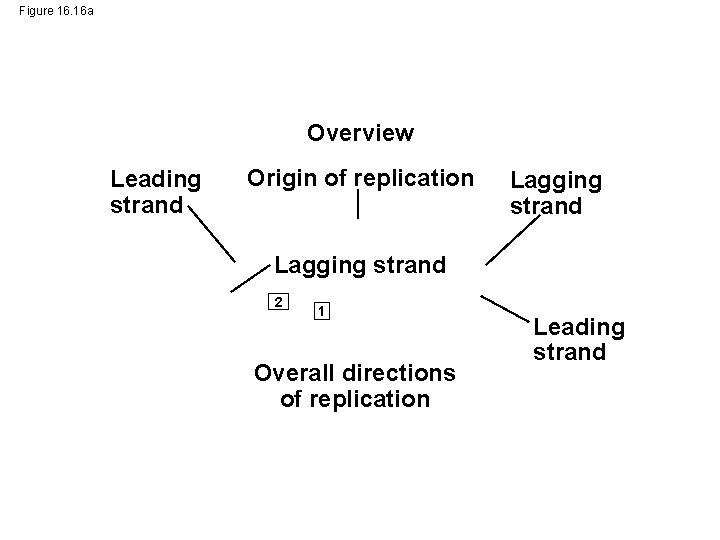
Figure 16. 16 a Overview Leading strand Origin of replication Lagging strand 2 1 Overall directions of replication Leading strand

Figure 16. 16 b-1 3 Template strand 5 3 5

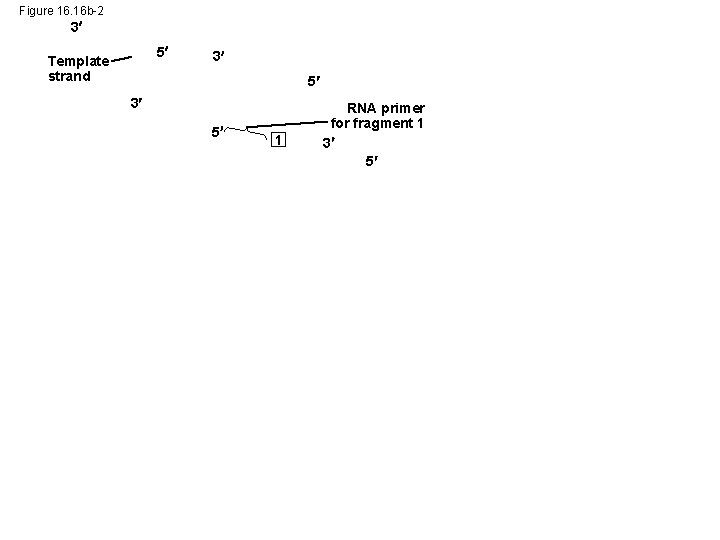
Figure 16. 16 b-2 3 5 Template strand 3 5 1 RNA primer for fragment 1 3 5

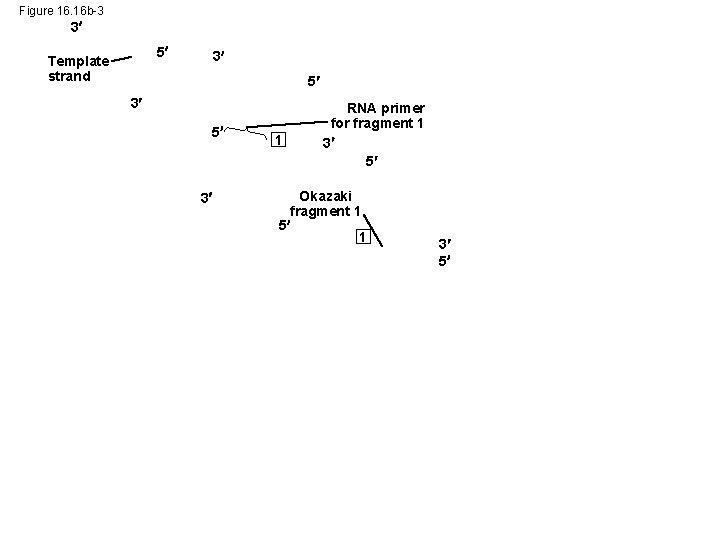
Figure 16. 16 b-3 3 5 Template strand 3 5 3 1 RNA primer for fragment 1 3 5 Okazaki fragment 1 5 1 3 5

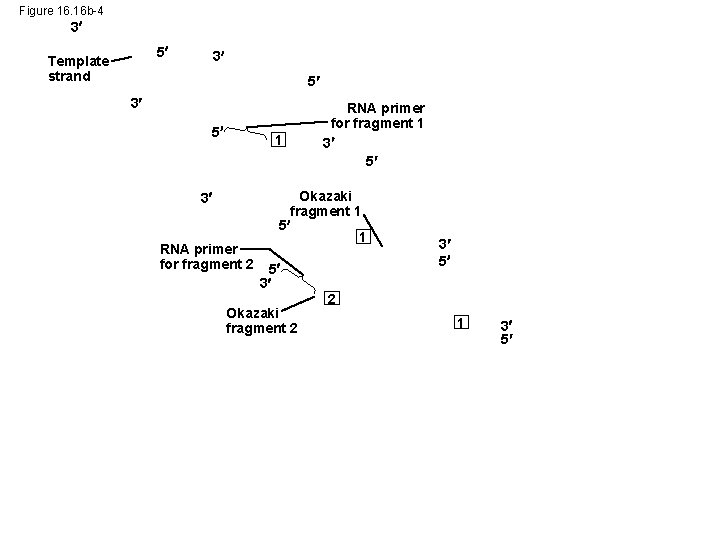
Figure 16. 16 b-4 3 5 Template strand 3 5 1 3 RNA primer for fragment 2 RNA primer for fragment 1 3 5 Okazaki fragment 1 5 3 Okazaki fragment 2 3 5 2 1 3 5

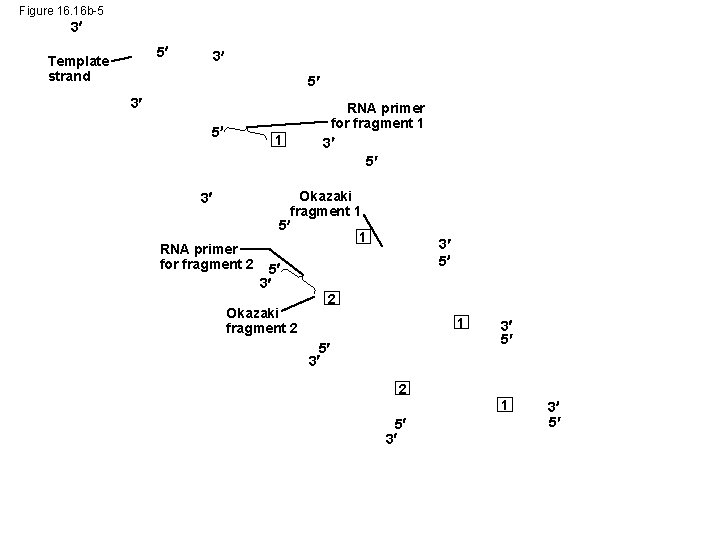
Figure 16. 16 b-5 3 5 Template strand 3 5 1 3 RNA primer for fragment 2 RNA primer for fragment 1 3 5 Okazaki fragment 1 5 3 Okazaki fragment 2 3 5 2 1 5 3 3 5

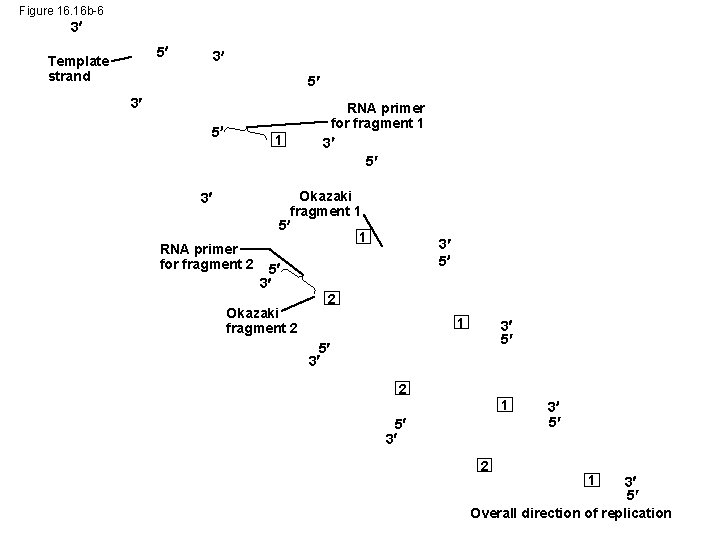
Figure 16. 16 b-6 3 5 Template strand 3 5 1 3 RNA primer for fragment 2 RNA primer for fragment 1 3 5 Okazaki fragment 1 5 3 Okazaki fragment 2 3 5 2 1 3 5 5 3 2 1 5 3 2 3 5 1 3 5 Overall direction of replication

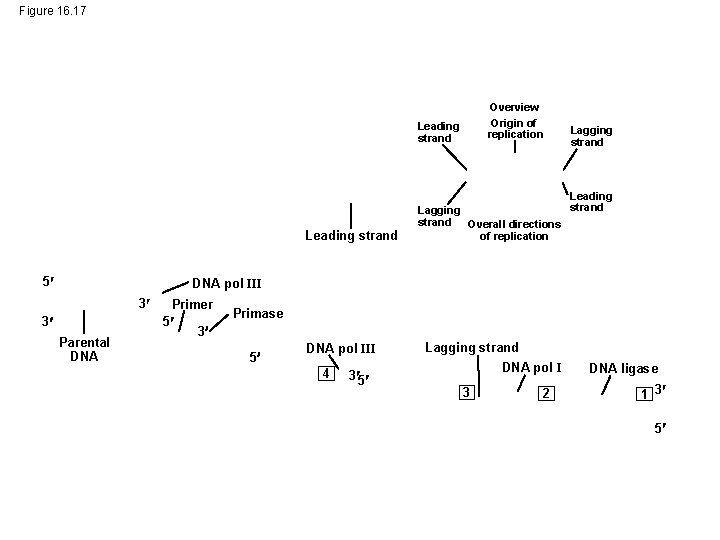
Figure 16. 17 Overview Origin of replication Leading strand 5 Lagging strand Overall directions of replication Lagging strand Leading strand DNA pol III 3 3 Parental DNA Primer 5 3 Primase 5 DNA pol III 4 3 5 Lagging strand DNA pol I 3 2 DNA ligase 1 3 5

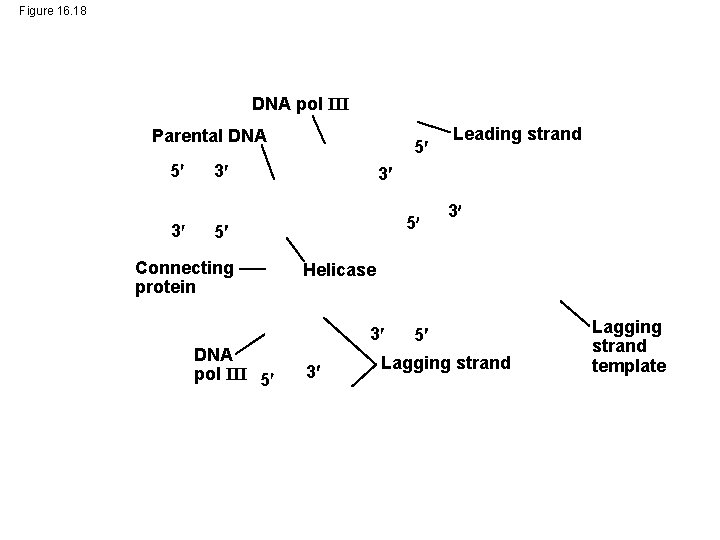
Figure 16. 18 DNA pol III Parental DNA 5 3 5 3 3 5 5 Connecting protein 3 Helicase 3 DNA pol III 5 Leading strand 3 5 Lagging strand template

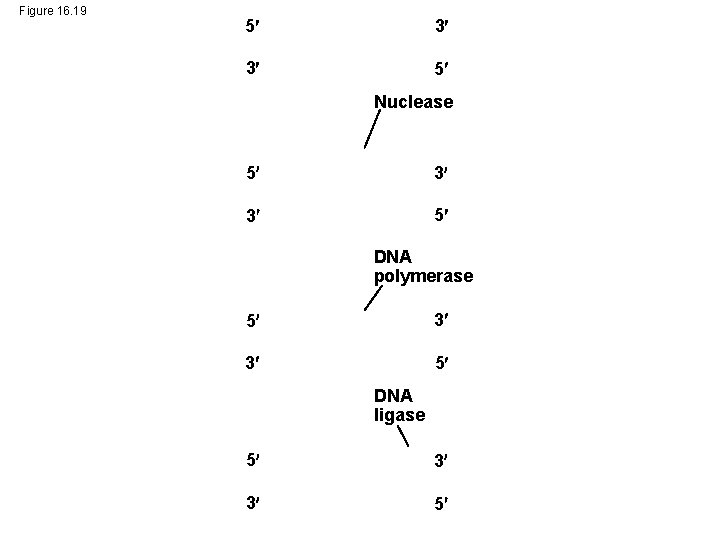
Figure 16. 19 5 3 3 5 Nuclease 5 3 3 5 DNA polymerase 5 3 3 5 DNA ligase 5 3 3 5

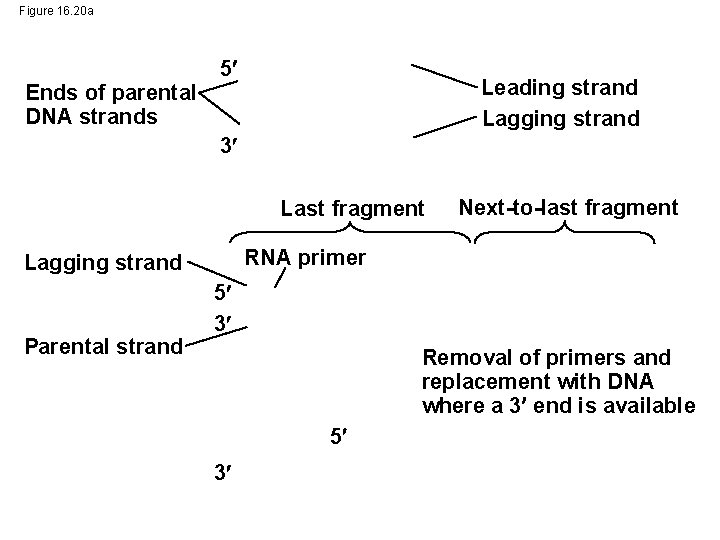
Figure 16. 20 a Ends of parental DNA strands 5 Leading strand Lagging strand 3 Last fragment RNA primer Lagging strand Parental strand Next-to-last fragment 5 3 Removal of primers and replacement with DNA where a 3 end is available 5 3

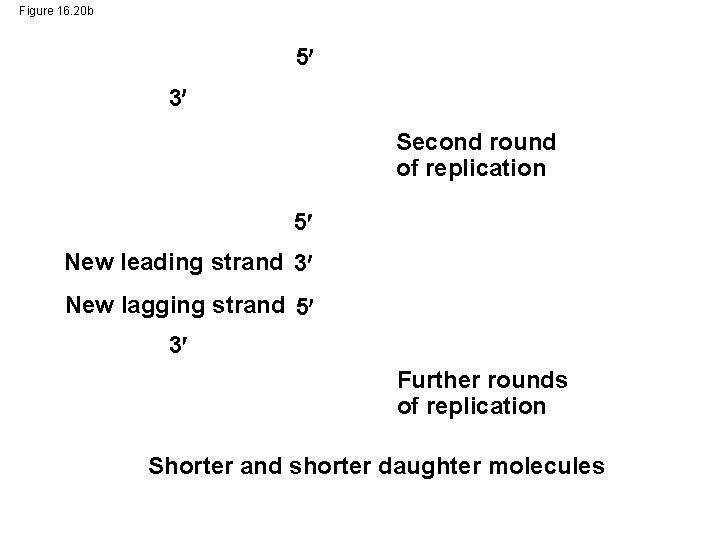
Figure 16. 20 b 5 3 Second round of replication 5 New leading strand 3 New lagging strand 5 3 Further rounds of replication Shorter and shorter daughter molecules

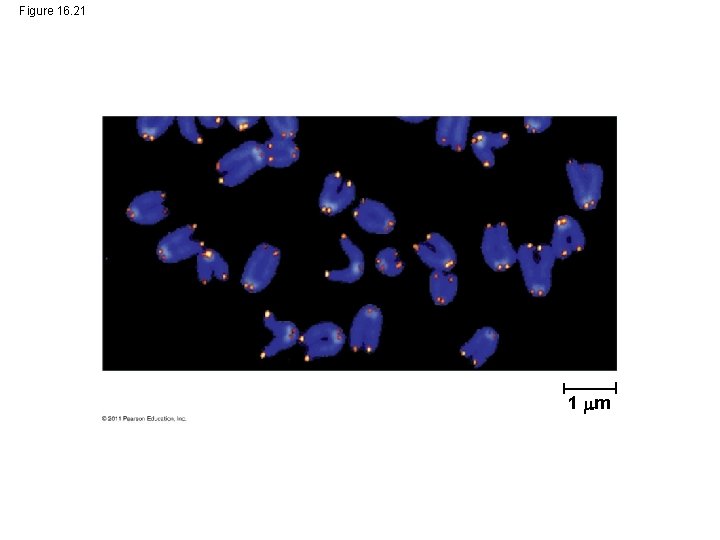
Figure 16. 21 1 m

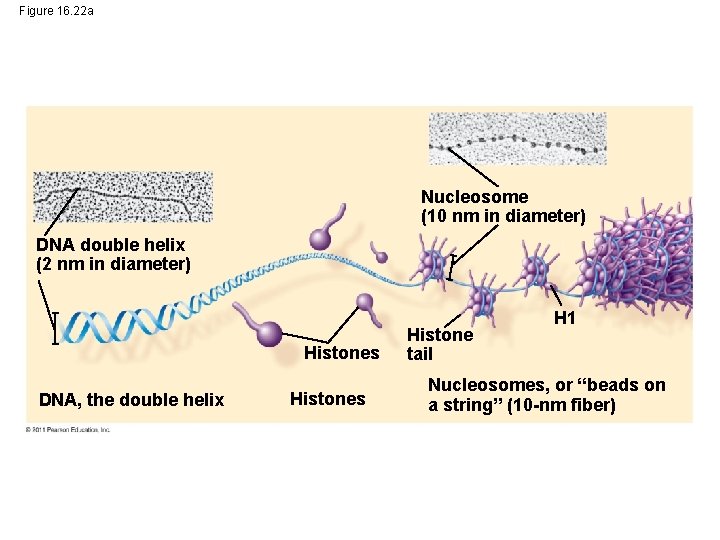
Figure 16. 22 a Nucleosome (10 nm in diameter) DNA double helix (2 nm in diameter) Histones DNA, the double helix Histones Histone tail H 1 Nucleosomes, or “beads on a string” (10 -nm fiber)

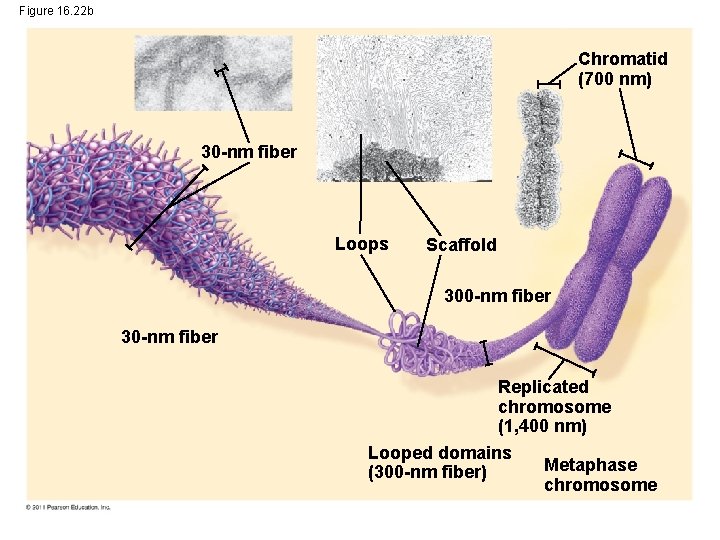
Figure 16. 22 b Chromatid (700 nm) 30 -nm fiber Loops Scaffold 300 -nm fiber 30 -nm fiber Replicated chromosome (1, 400 nm) Looped domains Metaphase (300 -nm fiber) chromosome

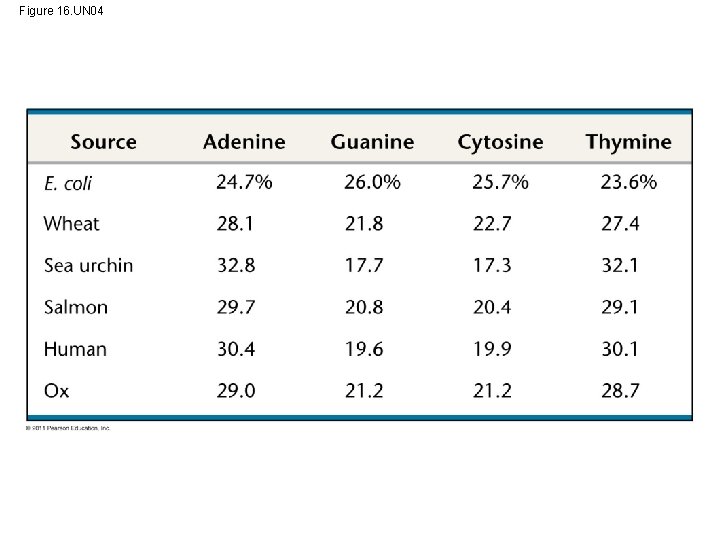
Figure 16. UN 04