Fig 18 8 1 A Eukaryotic Gene Enhancer
- Slides: 29

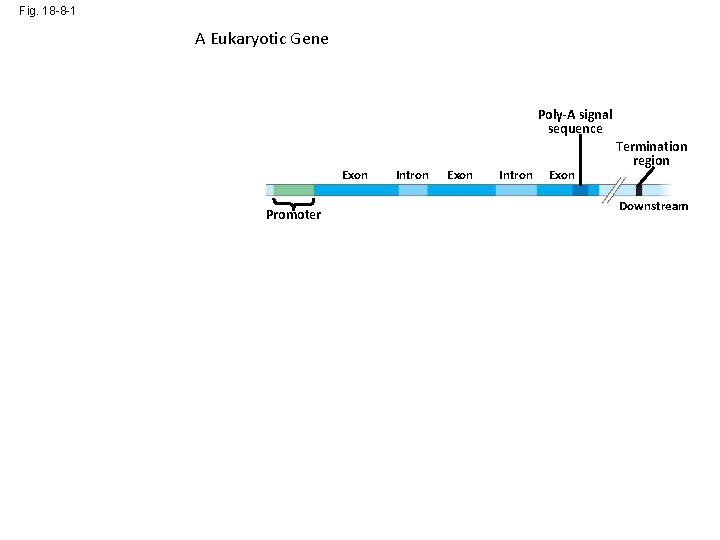
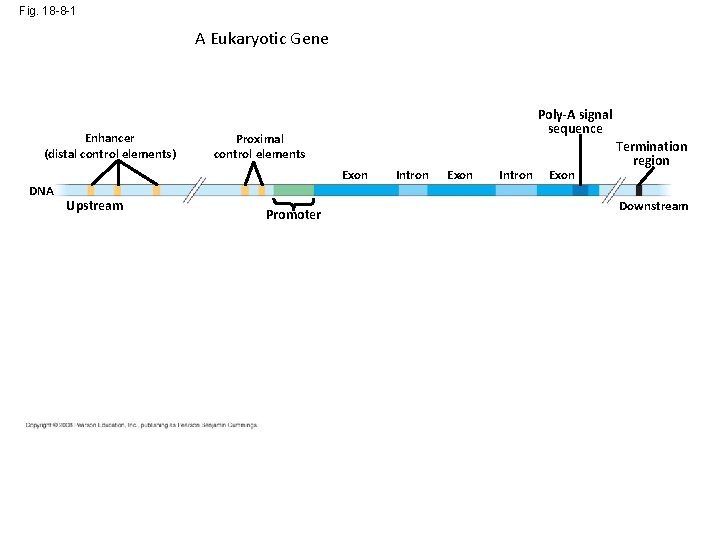
Fig. 18 -8 -1 A Eukaryotic Gene Enhancer (distal control elements) DNA Poly-A signal sequence Proximal control elements Exon Upstream Promoter Intron Exon Termination region Downstream

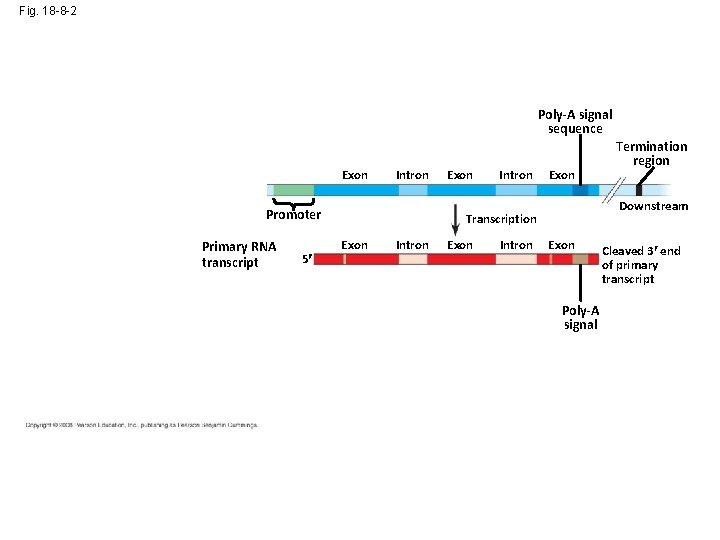
Fig. 18 -8 -2 Poly-A signal sequence Enhancer (distal control elements) DNA Exon Upstream Intron Promoter Primary RNA transcript 5 Exon Intron Exon Downstream Transcription Exon Intron Termination region Exon Poly-A signal Cleaved 3 end of primary transcript

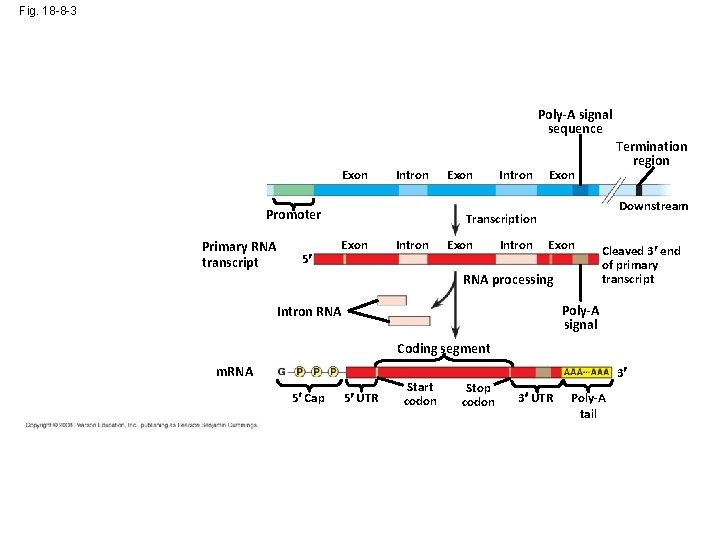
Fig. 18 -8 -3 Poly-A signal sequence Enhancer (distal control elements) DNA Exon Upstream Intron Promoter Primary RNA transcript 5 Exon Intron Termination region Exon Downstream Transcription Exon Intron Exon RNA processing Cleaved 3 end of primary transcript Poly-A signal Intron RNA Coding segment m. RNA 5 Cap 5 UTR Start codon Stop codon 3 3 UTR Poly-A tail

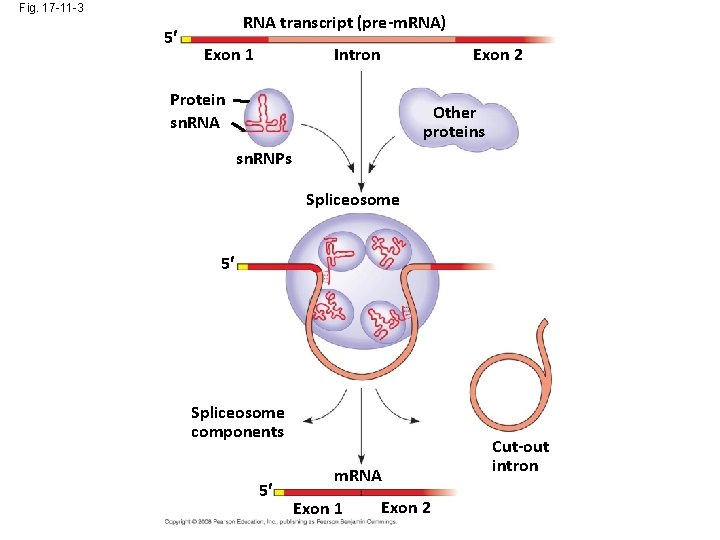
Fig. 17 -11 -3 5 RNA transcript (pre-m. RNA) Exon 1 Intron Exon 2 Protein sn. RNA Other proteins sn. RNPs Spliceosome 5 Spliceosome components 5 m. RNA Exon 1 Exon 2 Cut-out intron

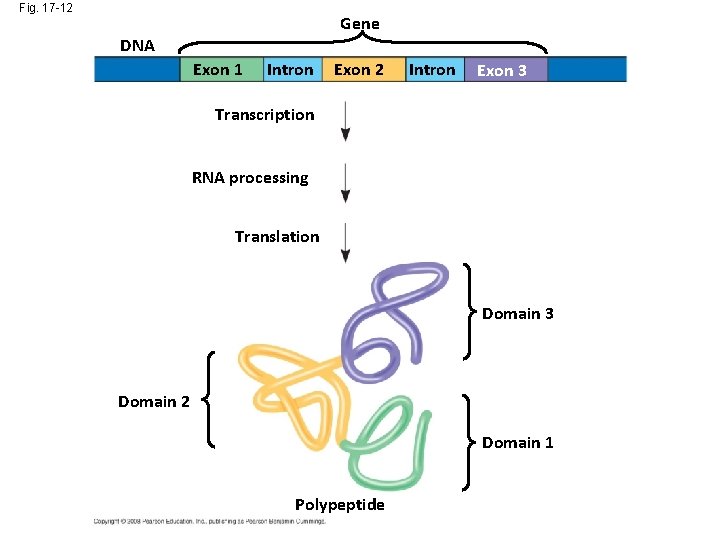
Fig. 17 -12 Gene DNA Exon 1 Intron Exon 2 Intron Exon 3 Transcription RNA processing Translation Domain 3 Domain 2 Domain 1 Polypeptide

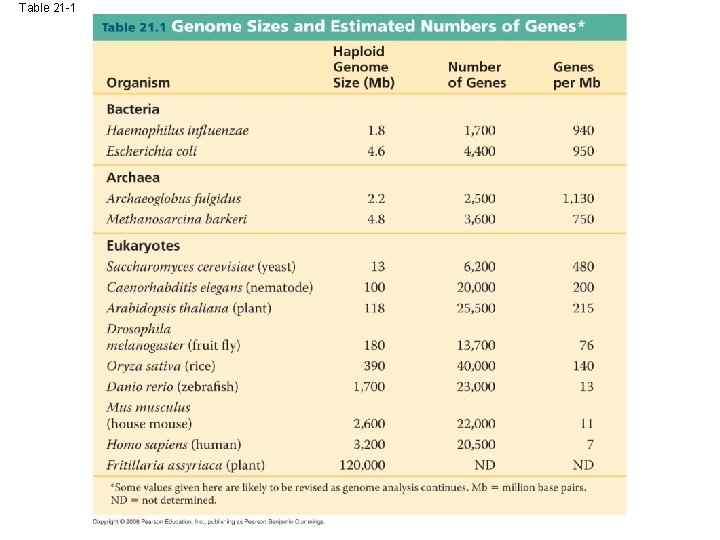
Table 21 -1

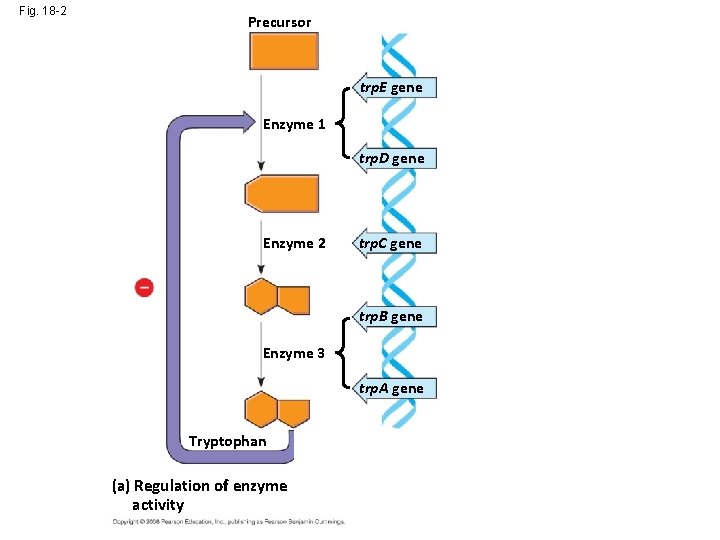
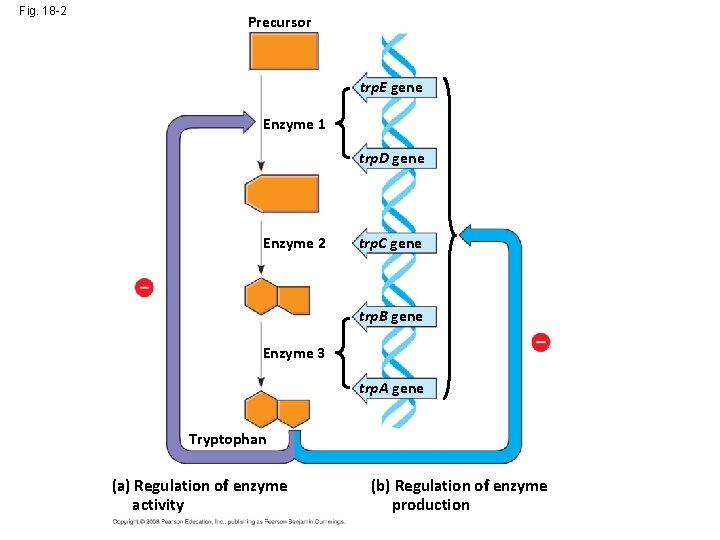
Fig. 18 -2 Precursor Feedback inhibition trp. E gene Enzyme 1 trp. D gene Regulation of gene expression Enzyme 2 trp. C gene trp. B gene Enzyme 3 trp. A gene Tryptophan (a) Regulation of enzyme activity (b) Regulation of enzyme production

Fig. 18 -2 Precursor trp. E gene Enzyme 1 trp. D gene Regulation of gene expression Enzyme 2 trp. C gene trp. B gene Enzyme 3 trp. A gene Tryptophan (a) Regulation of enzyme activity (b) Regulation of enzyme production

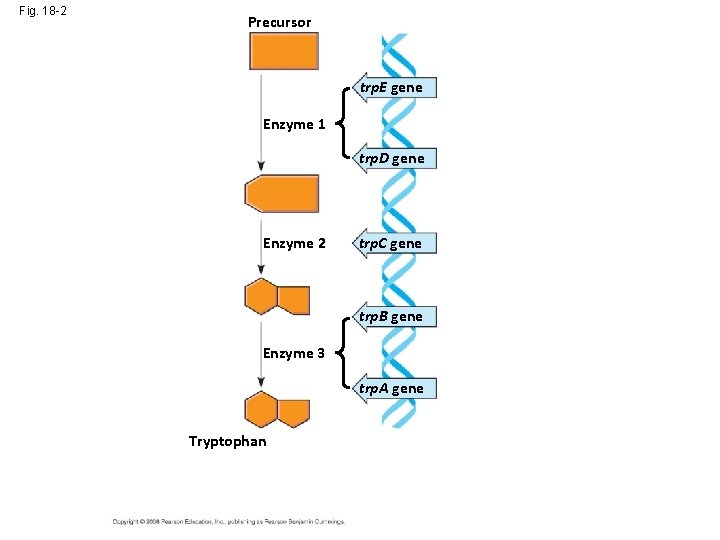
Fig. 18 -2 Precursor trp. E gene Enzyme 1 trp. D gene Enzyme 2 trp. C gene trp. B gene Enzyme 3 trp. A gene Tryptophan (a) Regulation of enzyme activity (b) Regulation of enzyme production

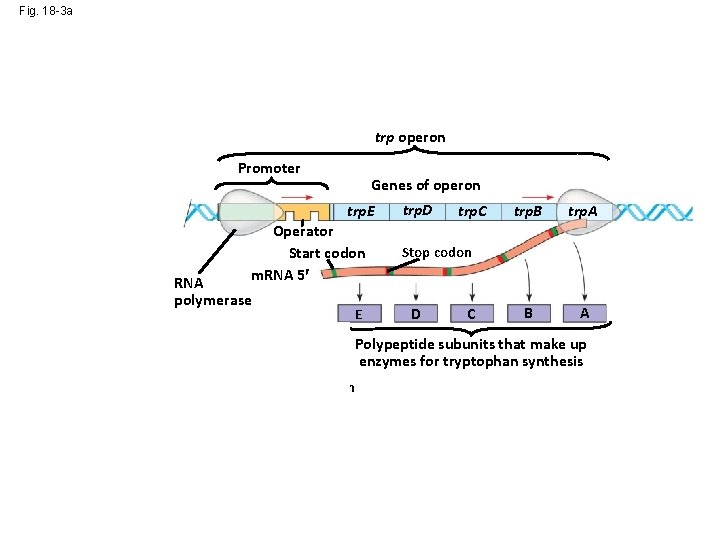
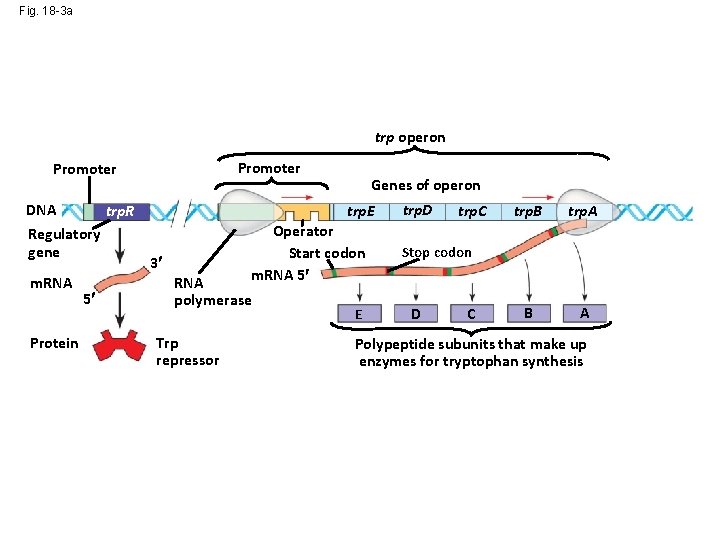
Fig. 18 -3 a trp operon Promoter DNA trp. R Regulatory gene m. RNA Protein 5 Genes of operon trp. E 3 Operator Start codon m. RNA 5 RNA polymerase Inactive repressor E trp. D trp. C trp. B trp. A B A Stop codon D C Polypeptide subunits that make up enzymes for tryptophan synthesis (a) Tryptophan absent, repressor inactive, operon on

Fig. 18 -3 a trp operon Promoter DNA trp. R Regulatory gene m. RNA Protein 5 Genes of operon trp. E 3 Operator Start codon m. RNA 5 RNA polymerase Trp repressor E trp. D trp. C trp. B trp. A B A Stop codon D C Polypeptide subunits that make up enzymes for tryptophan synthesis (a) Tryptophan absent, repressor inactive, operon on

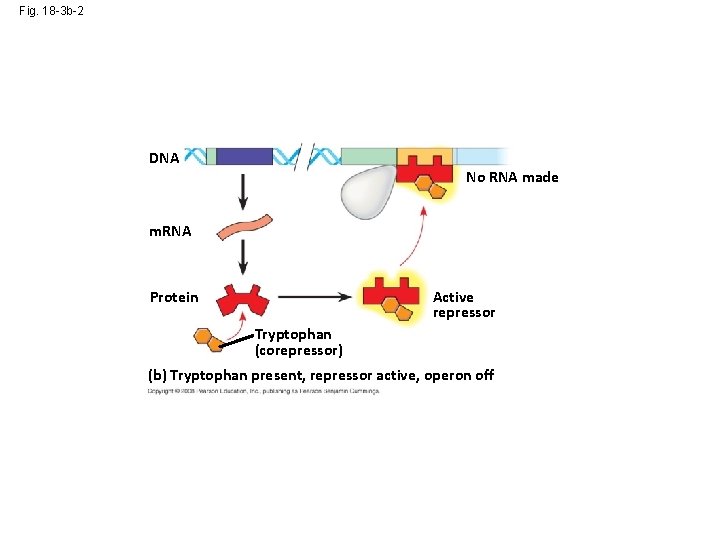
Fig. 18 -3 b-2 DNA No RNA made m. RNA Protein Active repressor Tryptophan (corepressor) (b) Tryptophan present, repressor active, operon off

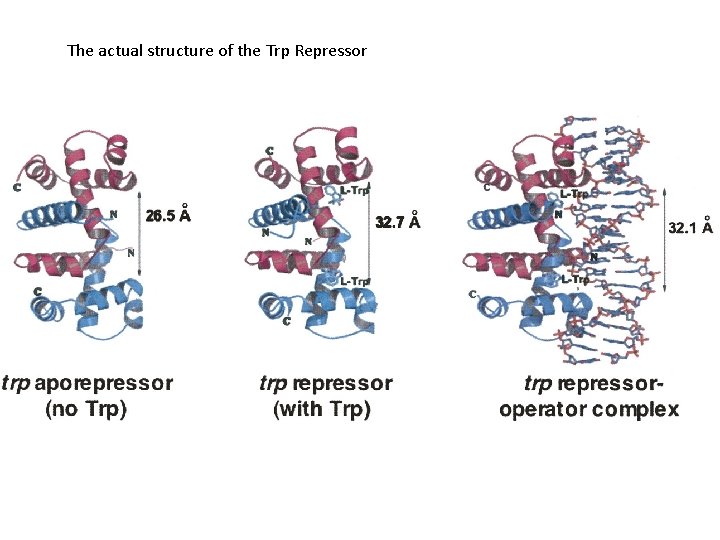
The actual structure of the Trp Repressor

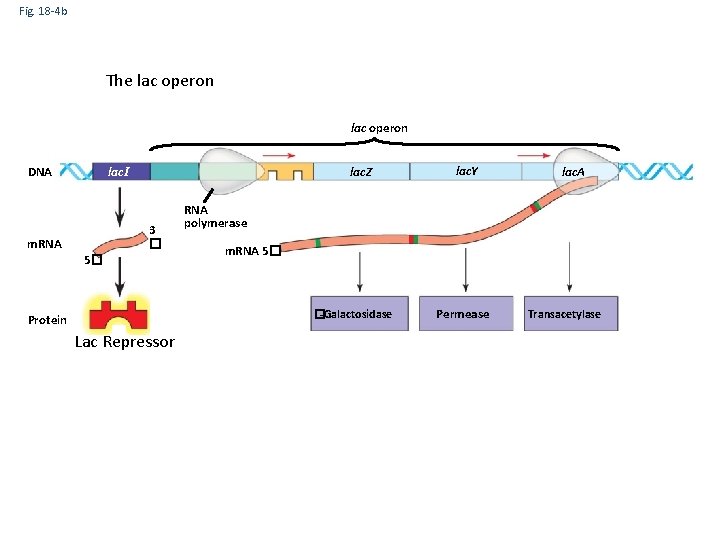
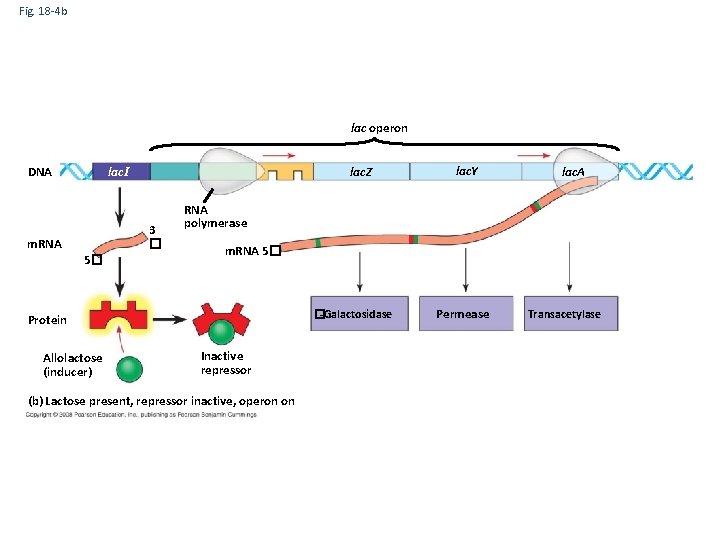
Fig. 18 -4 b The lac operon DNA lac. I lac. Z 3 � m. RNA 5� �-Galactosidase Allolactose (inducer) lac. A RNA polymerase Protein Lac Repressor lac. Y Inactive repressor (b) Lactose present, repressor inactive, operon on Permease Transacetylase

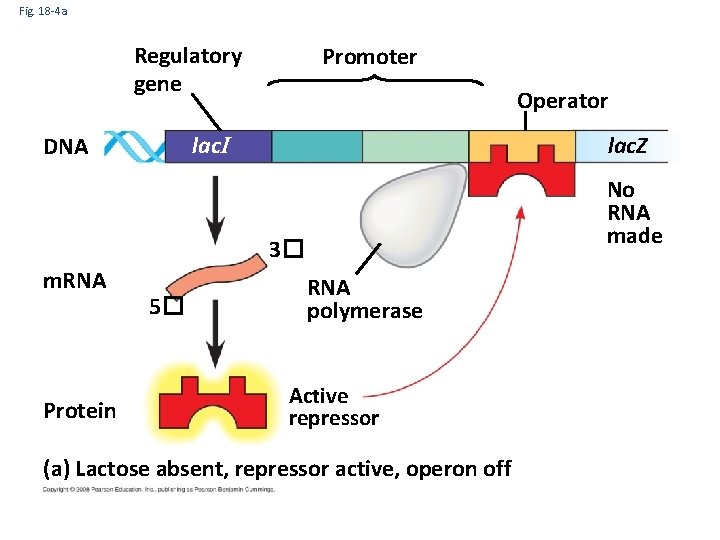
Fig. 18 -4 a Regulatory gene Promoter Operator lac. I DNA lac. Z No RNA made 3� m. RNA Protein 5� RNA polymerase Active repressor (a) Lactose absent, repressor active, operon off

Fig. 18 -4 b lac operon DNA lac. I lac. Z 3 � m. RNA 5� lac. A RNA polymerase m. RNA 5� �-Galactosidase Protein Allolactose (inducer) lac. Y Inactive repressor (b) Lactose present, repressor inactive, operon on Permease Transacetylase

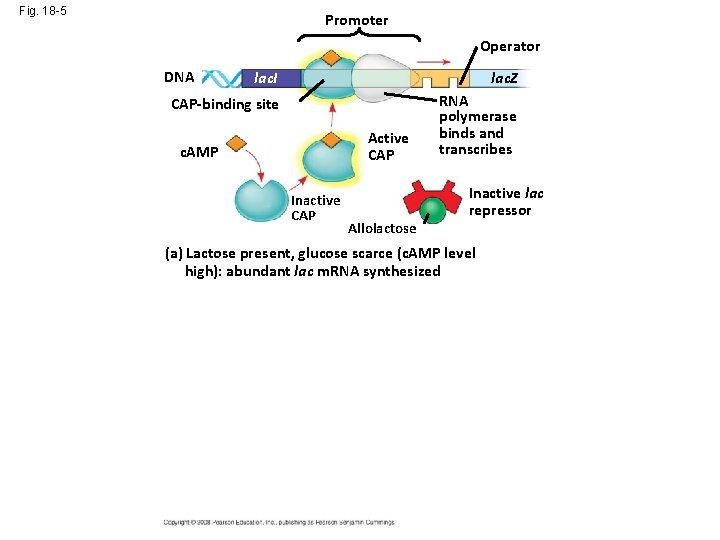
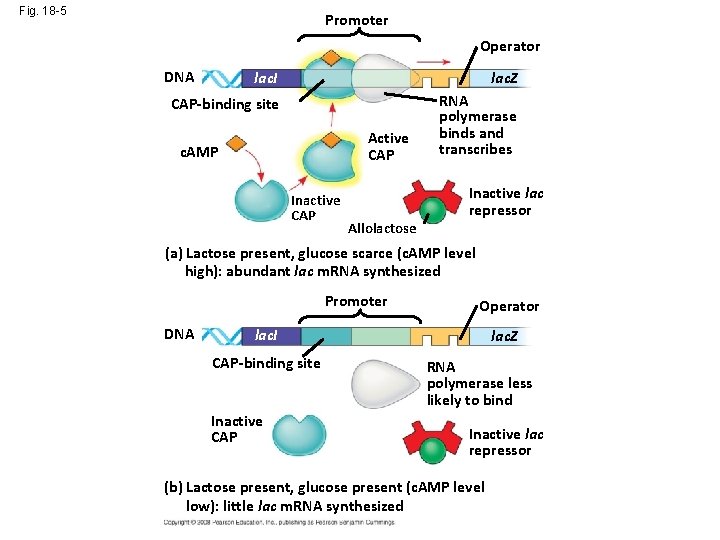
Fig. 18 -5 Promoter Operator DNA lac. I lac. Z CAP-binding site RNA polymerase binds and transcribes Active CAP c. AMP Inactive CAP Allolactose Inactive lac repressor (a) Lactose present, glucose scarce (c. AMP level high): abundant lac m. RNA synthesized Promoter DNA Operator lac. I CAP-binding site Inactive CAP lac. Z RNA polymerase less likely to bind Inactive lac repressor (b) Lactose present, glucose present (c. AMP level low): little lac m. RNA synthesized

Fig. 18 -5 Promoter Operator DNA lac. I lac. Z CAP-binding site RNA polymerase binds and transcribes Active CAP c. AMP Inactive CAP Allolactose Inactive lac repressor (a) Lactose present, glucose scarce (c. AMP level high): abundant lac m. RNA synthesized Promoter DNA Operator lac. I CAP-binding site Inactive CAP lac. Z RNA polymerase less likely to bind Inactive lac repressor (b) Lactose present, glucose present (c. AMP level low): little lac m. RNA synthesized

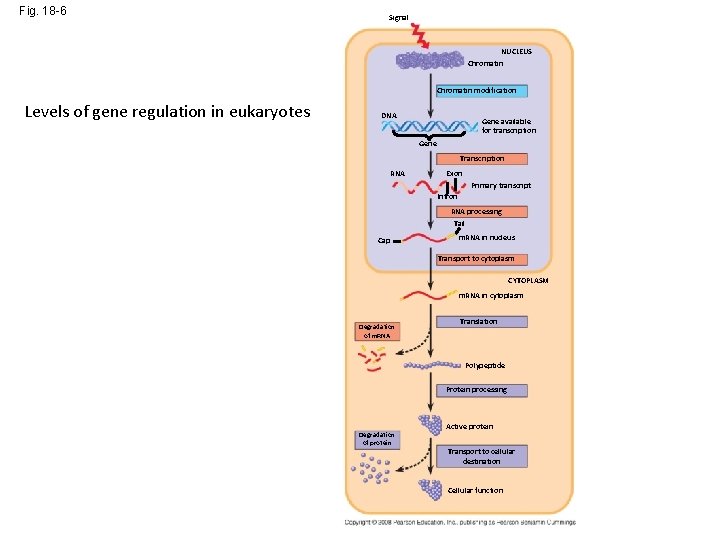
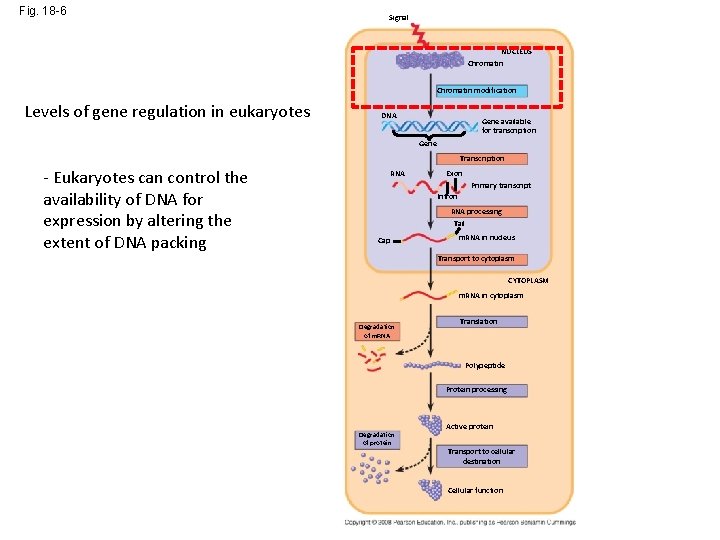
Fig. 18 -6 Signal NUCLEUS Chromatin modification Levels of gene regulation in eukaryotes DNA Gene available for transcription Gene Transcription RNA Exon Primary transcript Intron RNA processing Tail Cap m. RNA in nucleus Transport to cytoplasm CYTOPLASM m. RNA in cytoplasm Degradation of m. RNA Translation Polypeptide Protein processing Degradation of protein Active protein Transport to cellular destination Cellular function

Fig. 18 -6 Signal NUCLEUS Chromatin modification Levels of gene regulation in eukaryotes DNA Gene available for transcription Gene Transcription RNA Exon Primary transcript Intron RNA processing Tail Cap m. RNA in nucleus Transport to cytoplasm CYTOPLASM m. RNA in cytoplasm Degradation of m. RNA Translation Polypeptide Protein processing Degradation of protein Active protein Transport to cellular destination Cellular function

Fig. 18 -8 -1 A Eukaryotic Gene Enhancer (distal control elements) DNA Poly-A signal sequence Proximal control elements Exon Upstream Promoter Intron Exon Termination region Downstream

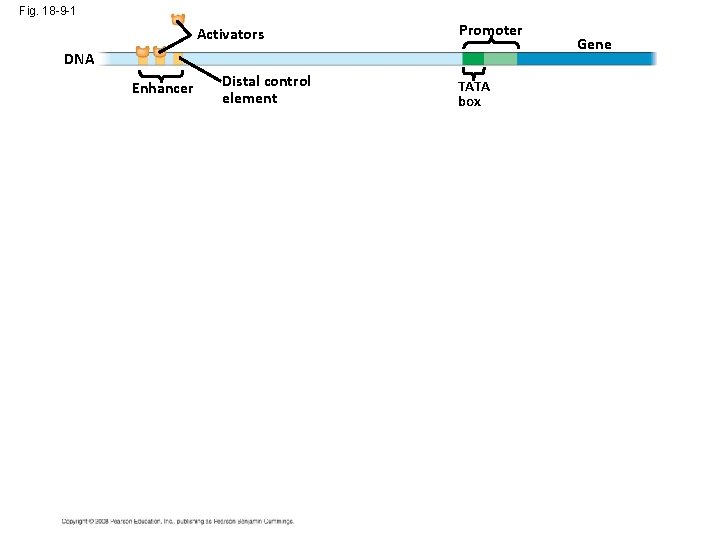
Fig. 18 -9 -1 Activators Promoter DNA Enhancer Distal control element TATA box Gene

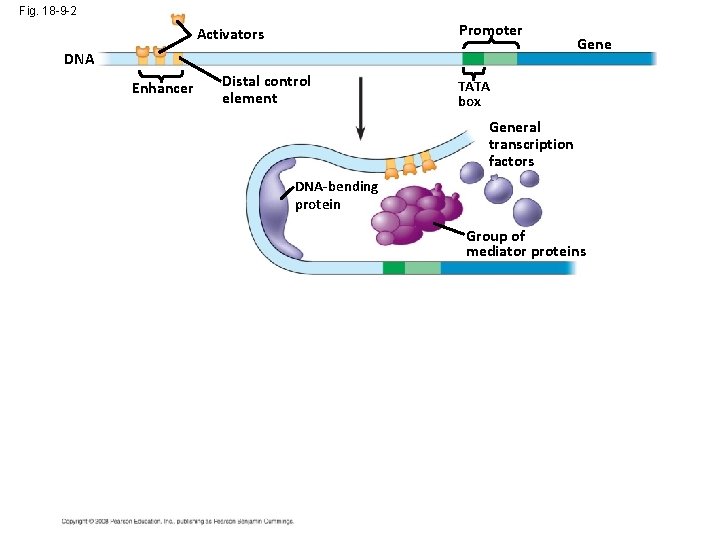
Fig. 18 -9 -2 Promoter Activators DNA Enhancer Distal control element Gene TATA box General transcription factors DNA-bending protein Group of mediator proteins

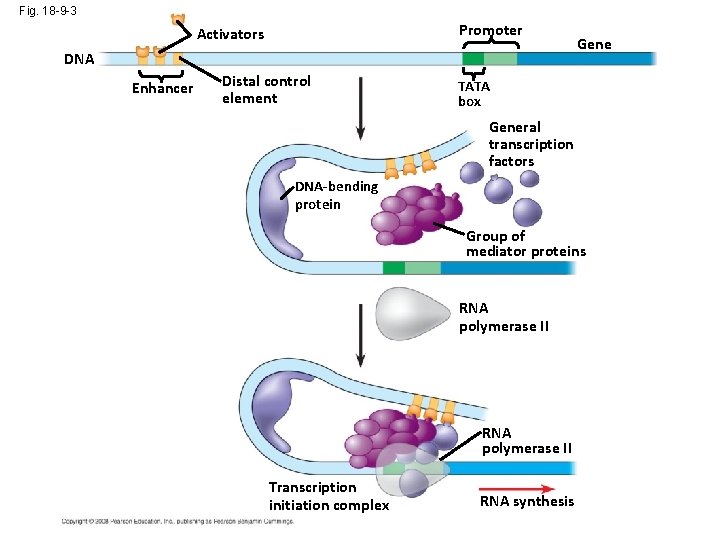
Fig. 18 -9 -3 Promoter Activators DNA Enhancer Distal control element Gene TATA box General transcription factors DNA-bending protein Group of mediator proteins RNA polymerase II Transcription initiation complex RNA synthesis

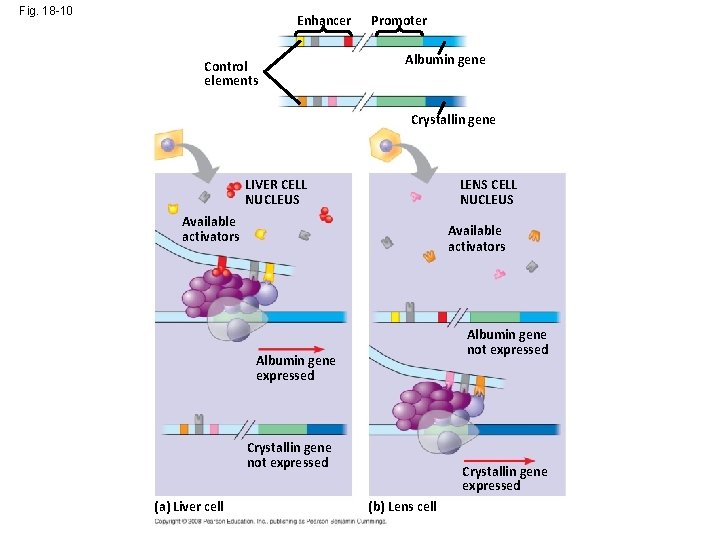
Fig. 18 -10 Enhancer Control elements Promoter Albumin gene Crystallin gene LIVER CELL NUCLEUS LENS CELL NUCLEUS Available activators Albumin gene not expressed Albumin gene expressed Crystallin gene not expressed (a) Liver cell Crystallin gene expressed (b) Lens cell

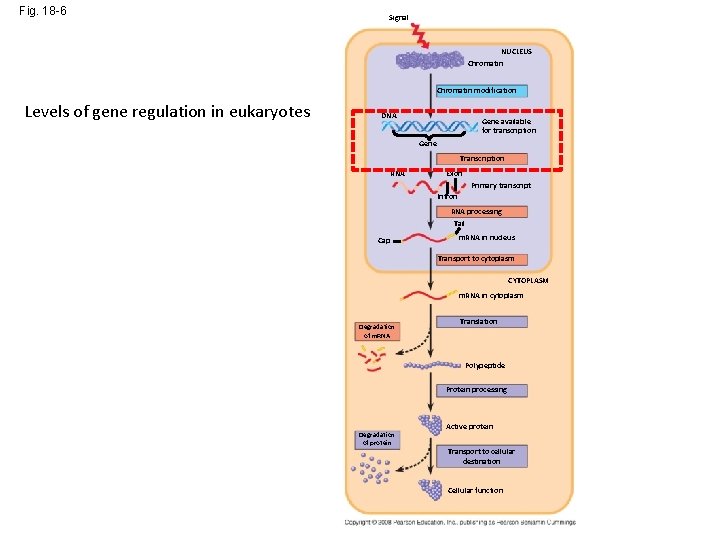
Fig. 18 -6 Signal NUCLEUS Chromatin modification Levels of gene regulation in eukaryotes DNA Gene available for transcription Gene Transcription - Eukaryotes can control the availability of DNA for expression by altering the extent of DNA packing RNA Exon Primary transcript Intron RNA processing Tail Cap m. RNA in nucleus Transport to cytoplasm CYTOPLASM m. RNA in cytoplasm Degradation of m. RNA Translation Polypeptide Protein processing Degradation of protein Active protein Transport to cellular destination Cellular function

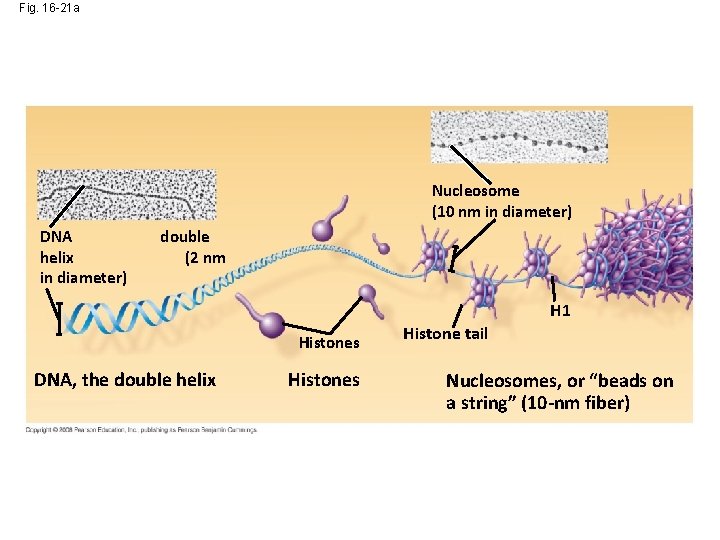
Fig. 16 -21 a Nucleosome (10 nm in diameter) DNA helix in diameter) double (2 nm H 1 Histones DNA, the double helix Histones Histone tail Nucleosomes, or “beads on a string” (10 -nm fiber)

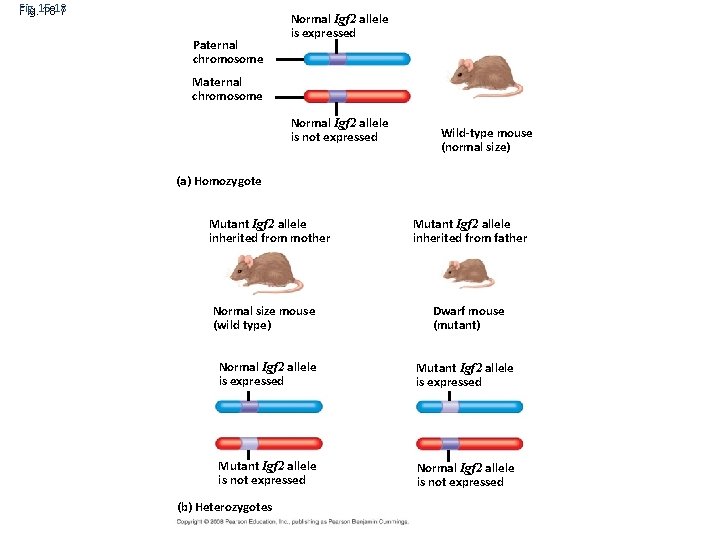
Fig. 15 -18 18 -7 Paternal chromosome Normal Igf 2 allele is expressed Maternal chromosome Normal Igf 2 allele is not expressed (a) Homozygote DNA double. Igf 2 helix Mutant allele inherited from mother Histone tails Wild-type mouse (normal size) Amino acids available for chemical modification Mutant Igf 2 allele inherited from father (a) Histone tails protrude outward from a nucleosome Normal size mouse Dwarf mouse (wild type) Normal Igf 2 allele is expressed Unacetylated histones (mutant) Mutant Igf 2 allele is expressed Acetylated histones Mutant Igf 2 allele Normal Igf 2 allele is not expressed (b) Acetylation of histone tails promotes loose chromatin structure that permits transcription (b) Heterozygotes

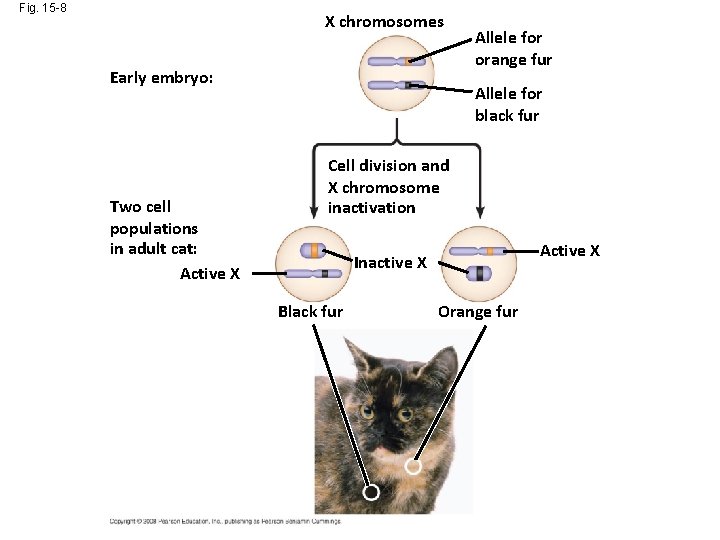
Fig. 15 -8 X chromosomes Early embryo: Two cell populations in adult cat: Active X Allele for orange fur Allele for black fur Cell division and X chromosome inactivation Active X Inactive X Black fur Orange fur